Eta categorical covariate plots (typical)
Usage
eta_vs_catcov(
xpdb,
mapping = NULL,
etavar = NULL,
drop_fixed = TRUE,
orientation = "x",
show_n = check_xpdb_x(xpdb, .warn = FALSE),
type = "bol",
title = "Eta versus categorical covariates | @run",
subtitle = "Based on @nind individuals, Eta shrink: @etashk",
caption = "@dir",
tag = NULL,
facets,
.problem,
quiet,
...
)Arguments
- xpdb
<
xp_xtras> or <xpose_data`> object- mapping
ggplot2style mapping- etavar
tidyselectforetavariables- drop_fixed
As in
xpose- orientation
Passed to
xplot_boxplot- show_n
Add "N=" to plot
- type
Passed to
xplot_boxplot- title
Plot title
- subtitle
Plot subtitle
- caption
Plot caption
- tag
Plot tag
- facets
Additional facets
- .problem
Problem number
- quiet
Silence output
- ...
Any additional aesthetics.
Details
The ability to show number per covariate level is inspired
by the package pmplots, but is implements here within
the xpose ecosystem for consistency.
Examples
# \donttest{
eta_vs_catcov(xpdb_x)
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Tidying data by ID, DOSE, AMT, SS, II ... and 23 more variables
#> Warning: attributes are not identical across measure variables; they will be dropped
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Tidying data by ID, DOSE, AMT, SS, II ... and 23 more variables
#> Warning: attributes are not identical across measure variables; they will be dropped
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Tidying data by ID, DOSE, AMT, SS, II ... and 23 more variables
#> Warning: attributes are not identical across measure variables; they will be dropped
#> [[1]]
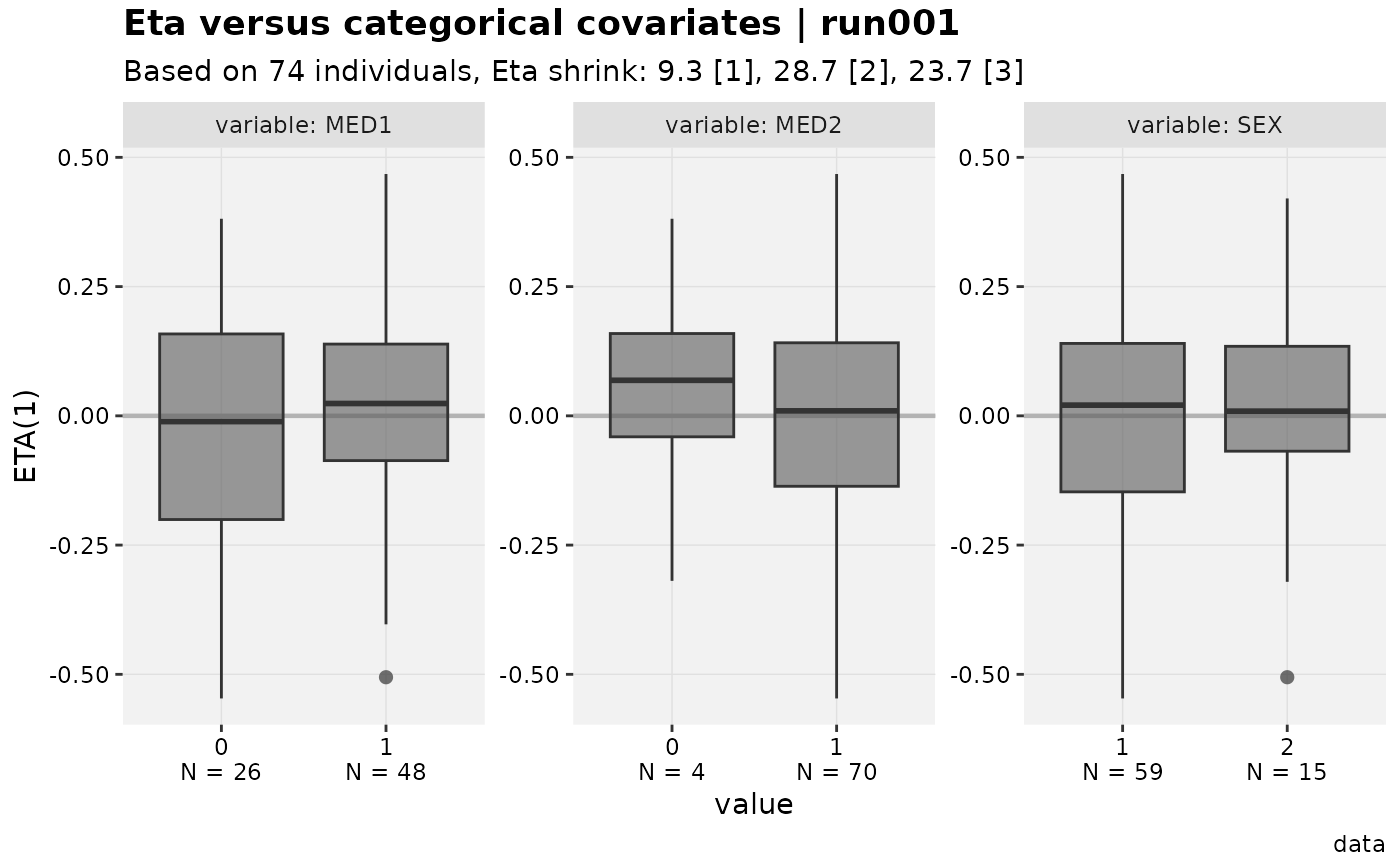 #>
#> [[2]]
#>
#> [[2]]
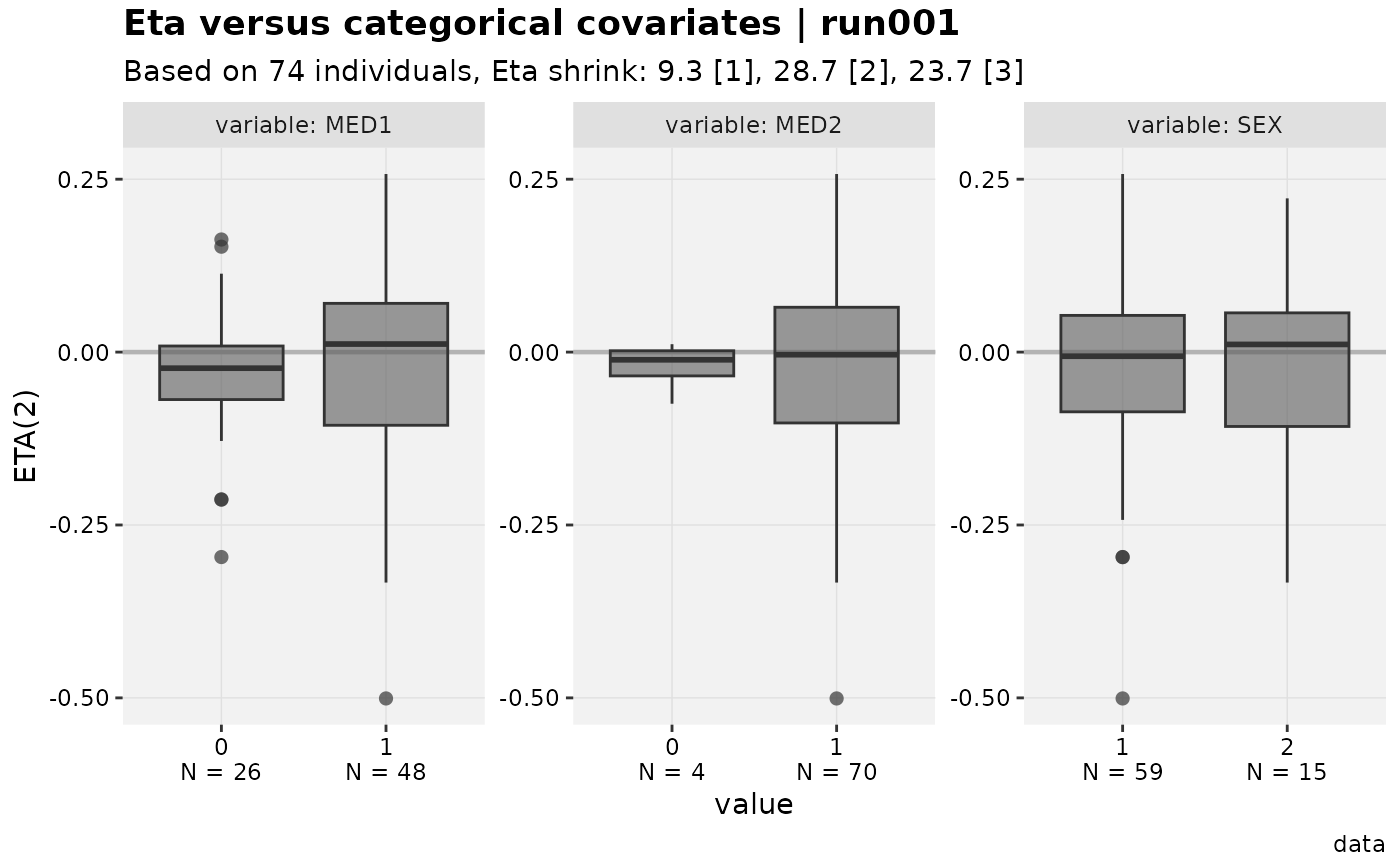 #>
#> [[3]]
#>
#> [[3]]
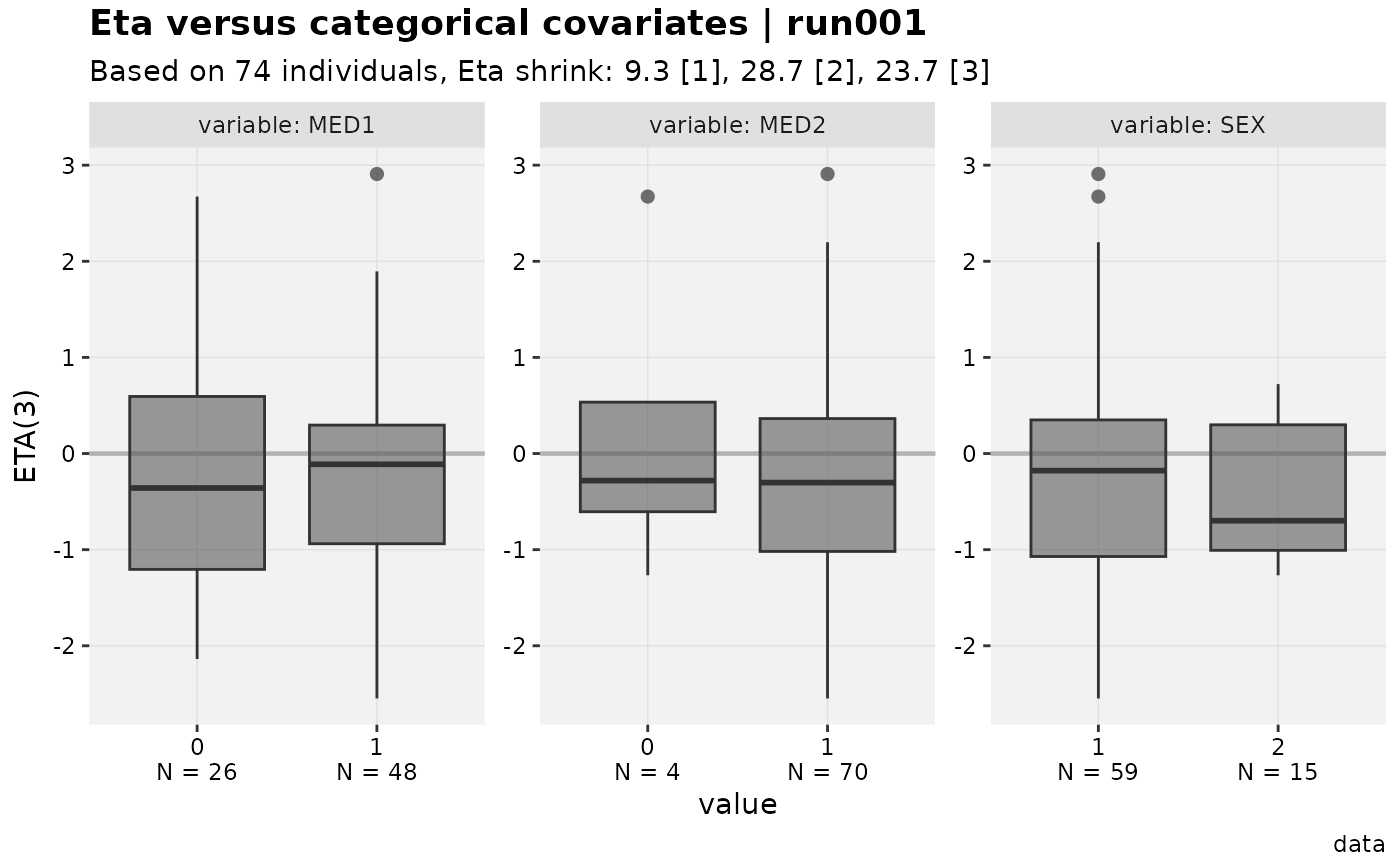 #>
# Labels and units are also supported
xpdb_x %>%
xpose::set_var_labels(AGE="Age", MED1 = "Digoxin") %>%
xpose::set_var_units(AGE="yrs") %>%
set_var_levels(SEX=lvl_sex(), MED1 = lvl_bin()) %>%
eta_vs_catcov()
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `out = purrr::map_if(...)`.
#> Caused by warning:
#> ! In $prob no.2 columns: MED1 not present in the data.
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Tidying data by ID, DOSE, AMT, SS, II ... and 23 more variables
#> Warning: attributes are not identical across measure variables; they will be dropped
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Tidying data by ID, DOSE, AMT, SS, II ... and 23 more variables
#> Warning: attributes are not identical across measure variables; they will be dropped
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Tidying data by ID, DOSE, AMT, SS, II ... and 23 more variables
#> Warning: attributes are not identical across measure variables; they will be dropped
#> [[1]]
#>
# Labels and units are also supported
xpdb_x %>%
xpose::set_var_labels(AGE="Age", MED1 = "Digoxin") %>%
xpose::set_var_units(AGE="yrs") %>%
set_var_levels(SEX=lvl_sex(), MED1 = lvl_bin()) %>%
eta_vs_catcov()
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `out = purrr::map_if(...)`.
#> Caused by warning:
#> ! In $prob no.2 columns: MED1 not present in the data.
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Tidying data by ID, DOSE, AMT, SS, II ... and 23 more variables
#> Warning: attributes are not identical across measure variables; they will be dropped
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Tidying data by ID, DOSE, AMT, SS, II ... and 23 more variables
#> Warning: attributes are not identical across measure variables; they will be dropped
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Tidying data by ID, DOSE, AMT, SS, II ... and 23 more variables
#> Warning: attributes are not identical across measure variables; they will be dropped
#> [[1]]
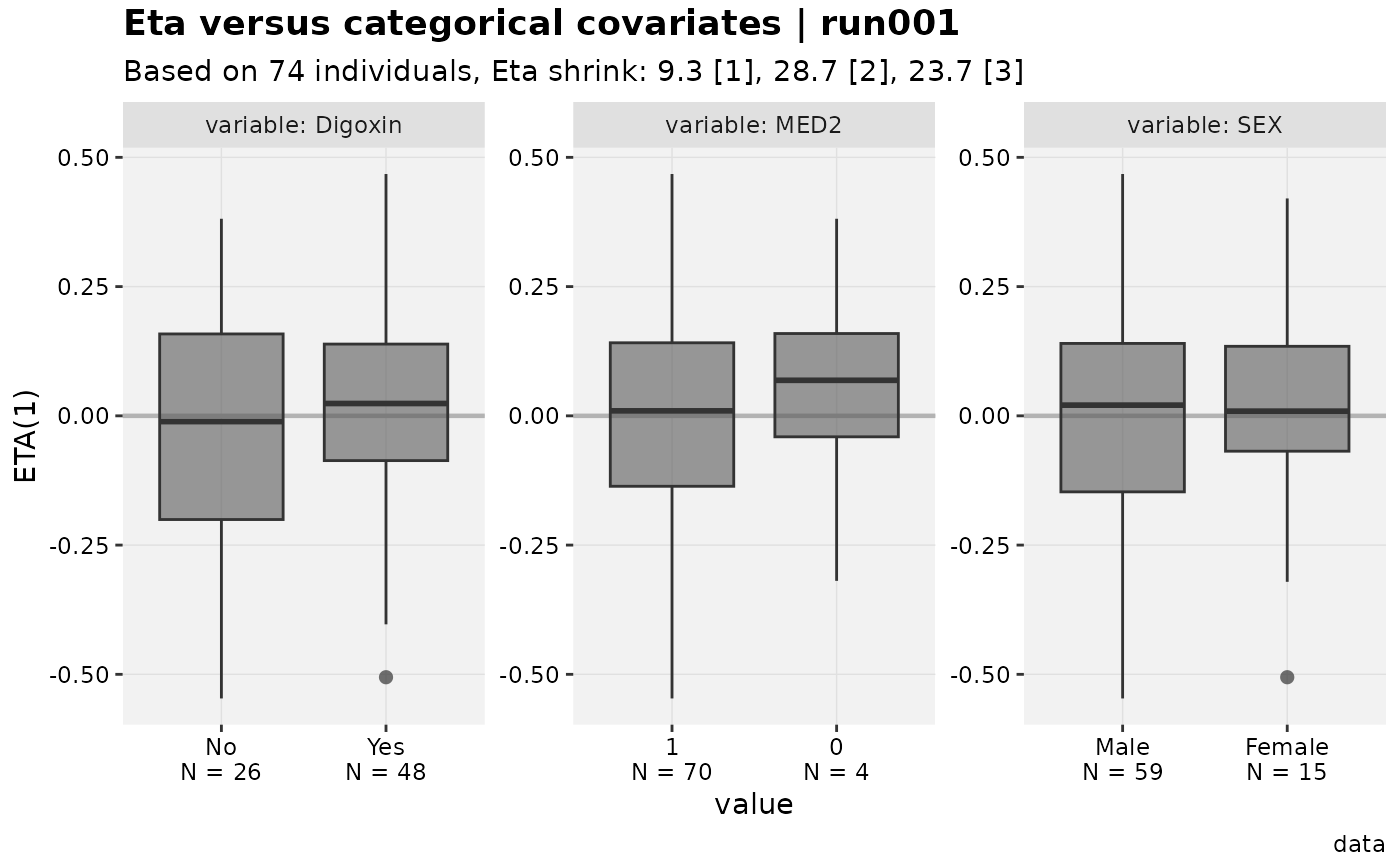 #>
#> [[2]]
#>
#> [[2]]
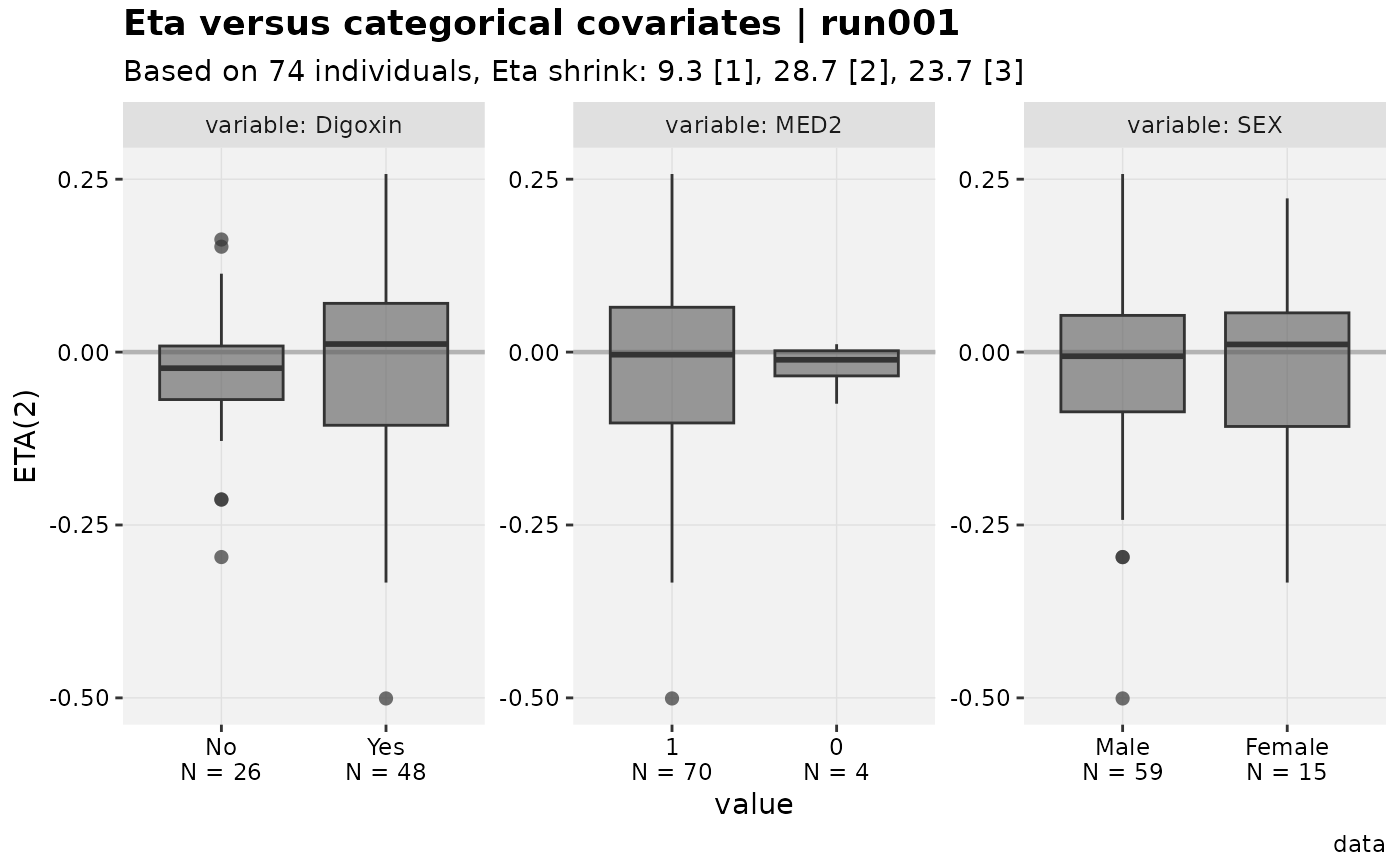 #>
#> [[3]]
#>
#> [[3]]
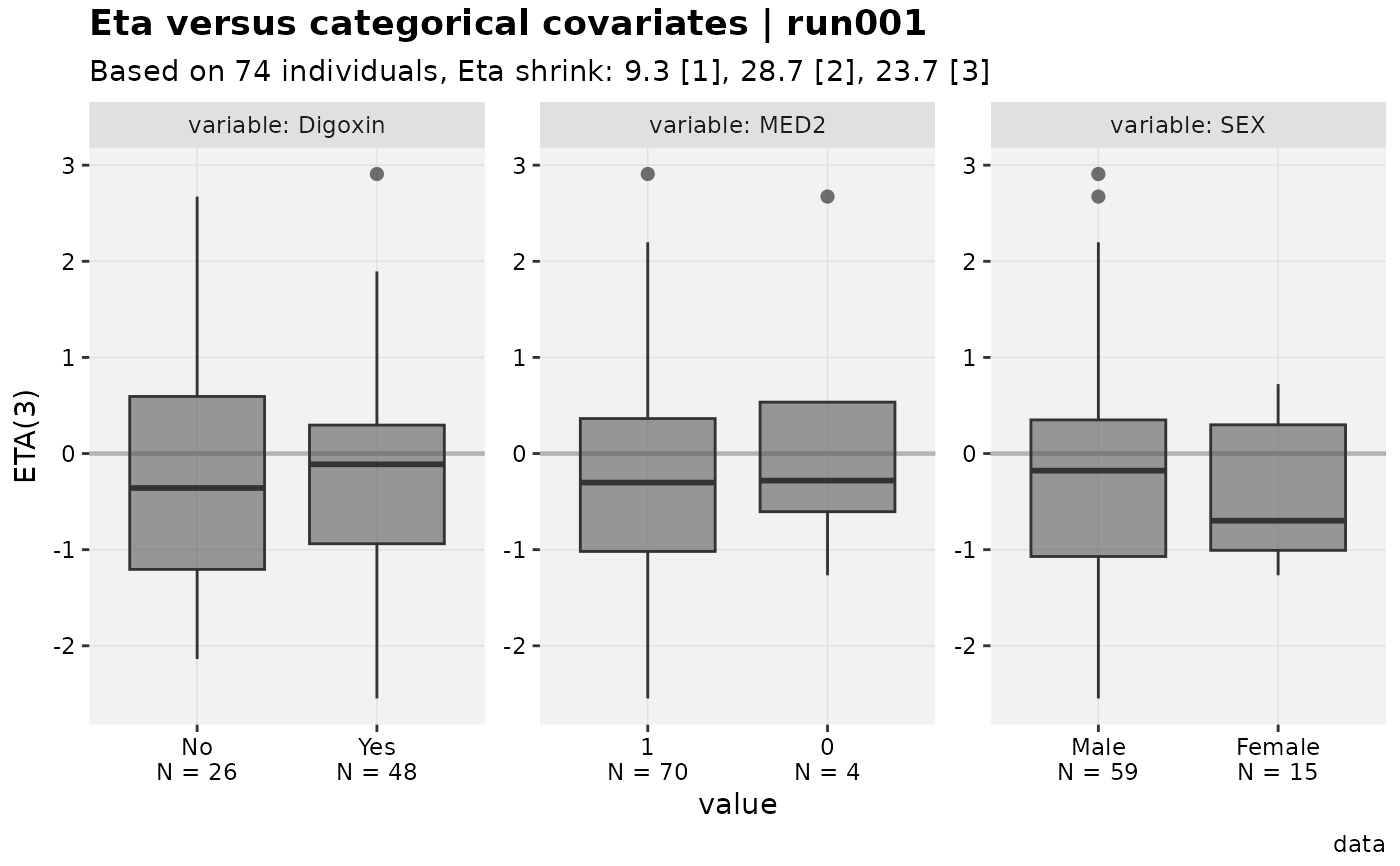 #>
# }
#>
# }
