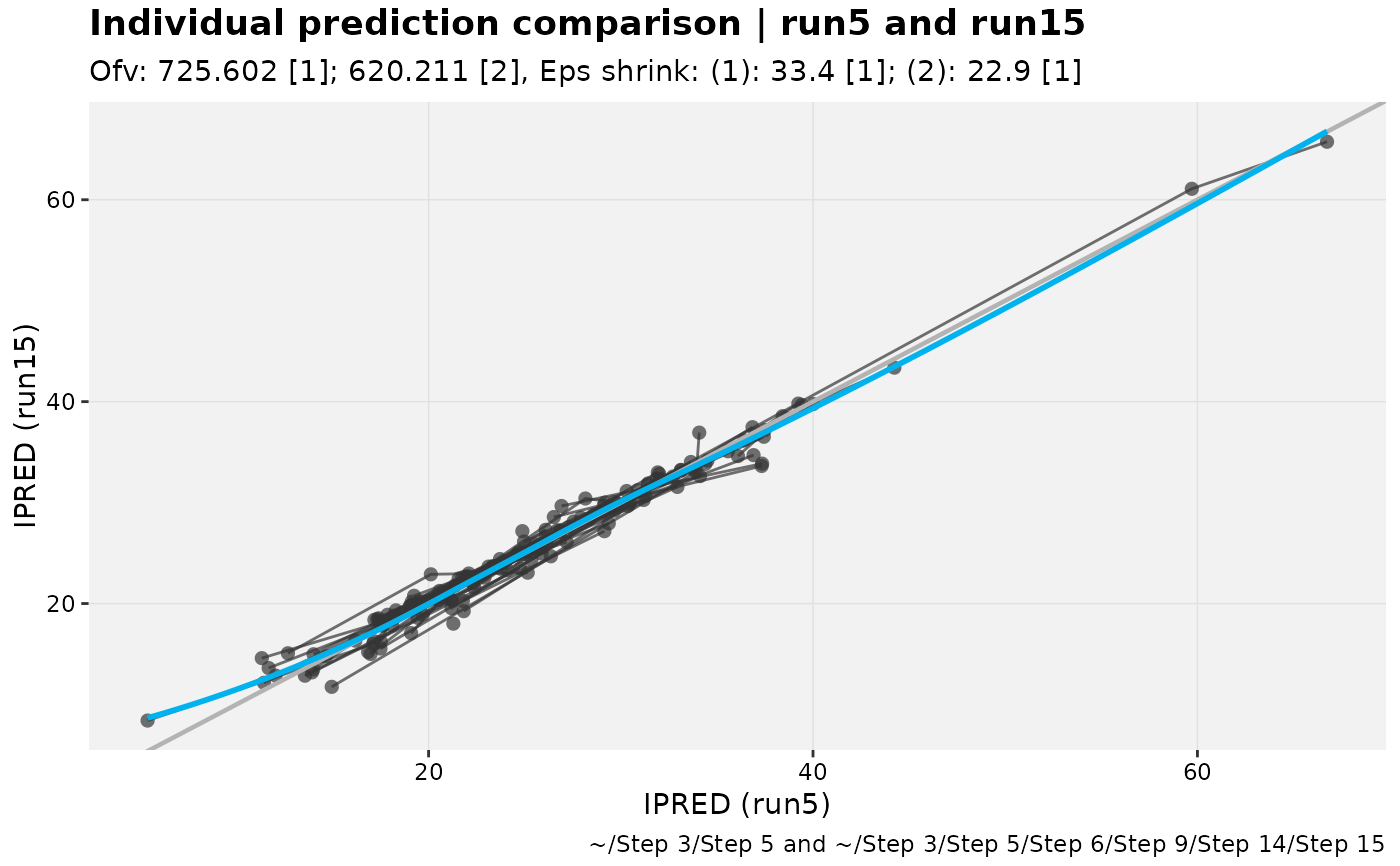
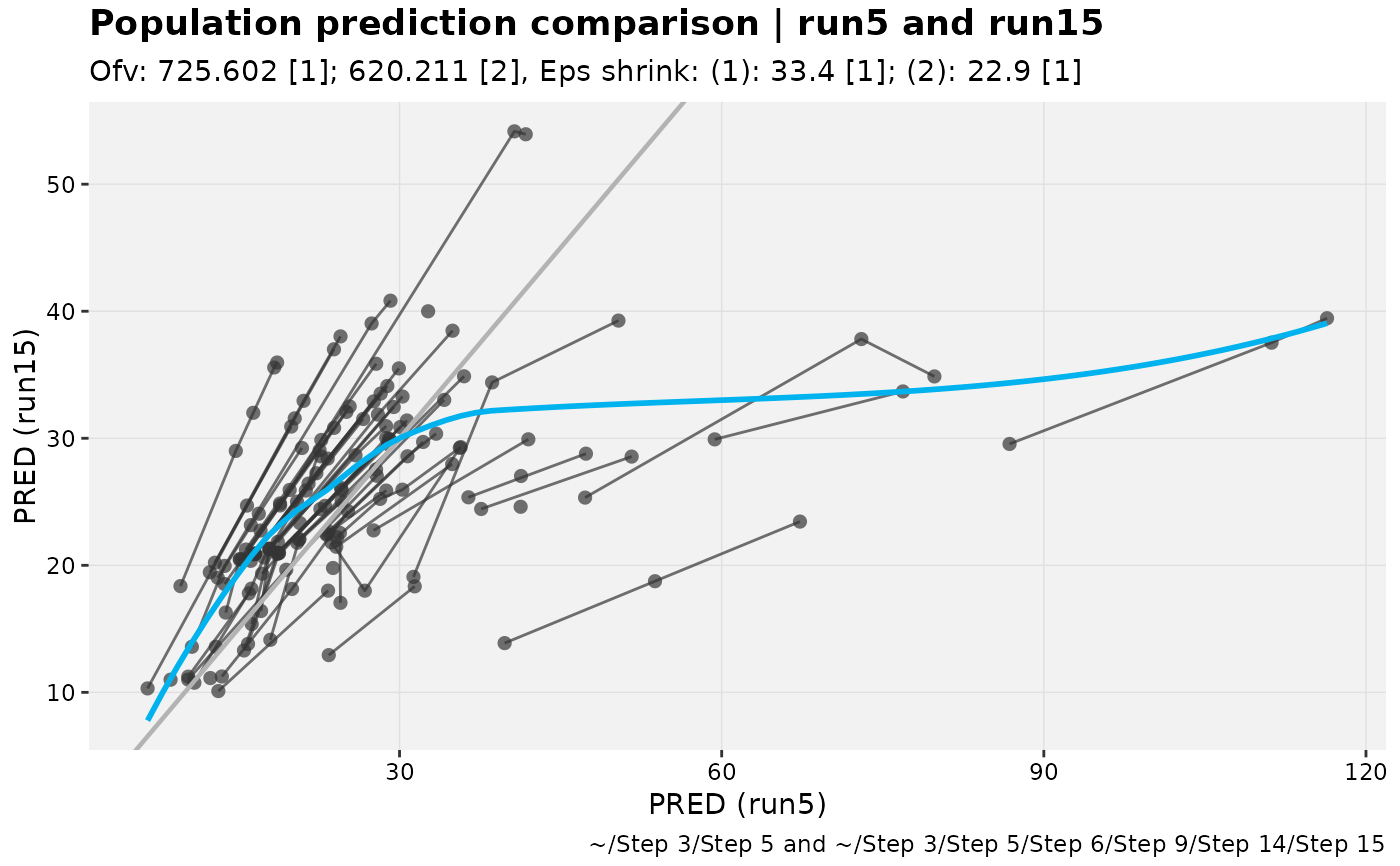
For two models in an xpose_set, these functions are useful in comparing individual
and population predictions
Usage
ipred_vs_ipred(
xpdb_s,
...,
.inorder = FALSE,
type = "pls",
title = "Individual prediction comparison | @run",
subtitle = "Ofv: @ofv, Eps shrink: @epsshk",
caption = "@dir",
tag = NULL,
log = NULL,
guide = TRUE,
axis.text = "@run",
facets,
.problem,
quiet
)
pred_vs_pred(
xpdb_s,
...,
.inorder = FALSE,
type = "pls",
title = "Population prediction comparison | @run",
subtitle = "Ofv: @ofv, Eps shrink: @epsshk",
caption = "@dir",
tag = NULL,
log = NULL,
guide = TRUE,
axis.text = "@run",
facets,
.problem,
quiet
)Arguments
- xpdb_s
<
xpose_set> object- ...
See <
two_set_dots>- .inorder
See <
two_set_dots>- type
Passed to
xplot_scatter- title
Plot title
- subtitle
Plot subtitle
- caption
Plot caption
- tag
Plot tag
- log
Log scale covariate value?
- guide
Add guide line?
- axis.text
What to show in the axes to distinguish the model values
- facets
Additional facets
- .problem
Problem number
- quiet
Silence output