This is essentially a wrapper around ggpairs,
except it uses xpose motifs and styling. Note that this function
produces a lot of repetitive output if quiet=FALSE; this may not
be an issue, but it could look like an error has occurred if many covariates
and individual parameter estimates are included.
Usage
eta_grid(
xpdb,
mapping = NULL,
etavar = NULL,
drop_fixed = TRUE,
title = "Eta correlations | @run",
subtitle = "Based on @nind individuals, Eta shrink: @etashk",
caption = "@dir",
tag = NULL,
pairs_opts,
.problem,
quiet,
...
)
cov_grid(
xpdb,
mapping = NULL,
cols = NULL,
covtypes = c("cont", "cat"),
show_n = TRUE,
drop_fixed = TRUE,
title = "Covariate relationships | @run",
subtitle = "Based on @nind individuals",
caption = "@dir",
tag = NULL,
pairs_opts,
.problem,
quiet,
...
)
eta_vs_cov_grid(
xpdb,
mapping = NULL,
etavar = NULL,
cols = NULL,
covtypes = c("cont", "cat"),
show_n = TRUE,
drop_fixed = TRUE,
title = "Eta covariate correlations | @run",
subtitle = "Based on @nind individuals, Eta shrink: @etashk",
caption = "@dir",
tag = NULL,
etacov = TRUE,
pairs_opts,
.problem,
quiet,
...
)Arguments
- xpdb
<
xp_xtras> or <xpose_data`> object- mapping
ggplot2style mapping- etavar
tidyselectforetavariables- drop_fixed
As in
xpose- title
Plot title
- subtitle
Plot subtitle
- caption
Plot caption
- tag
Plot tag
- pairs_opts
List of arguments to pass to
_opts. See <xplot_pairs>- .problem
Problem number
- quiet
Silence extra debugging output
- ...
Passed to
xplot_pairs- cols
tidyselectfor covariates variables- covtypes
Subset to specific covariate type?
- show_n
Count the number of
IDs in each category- etacov
For
eta_vs_cov_grid,etaare sorted after covariates to give anxorientation to covariate relationships.
Examples
# \donttest{
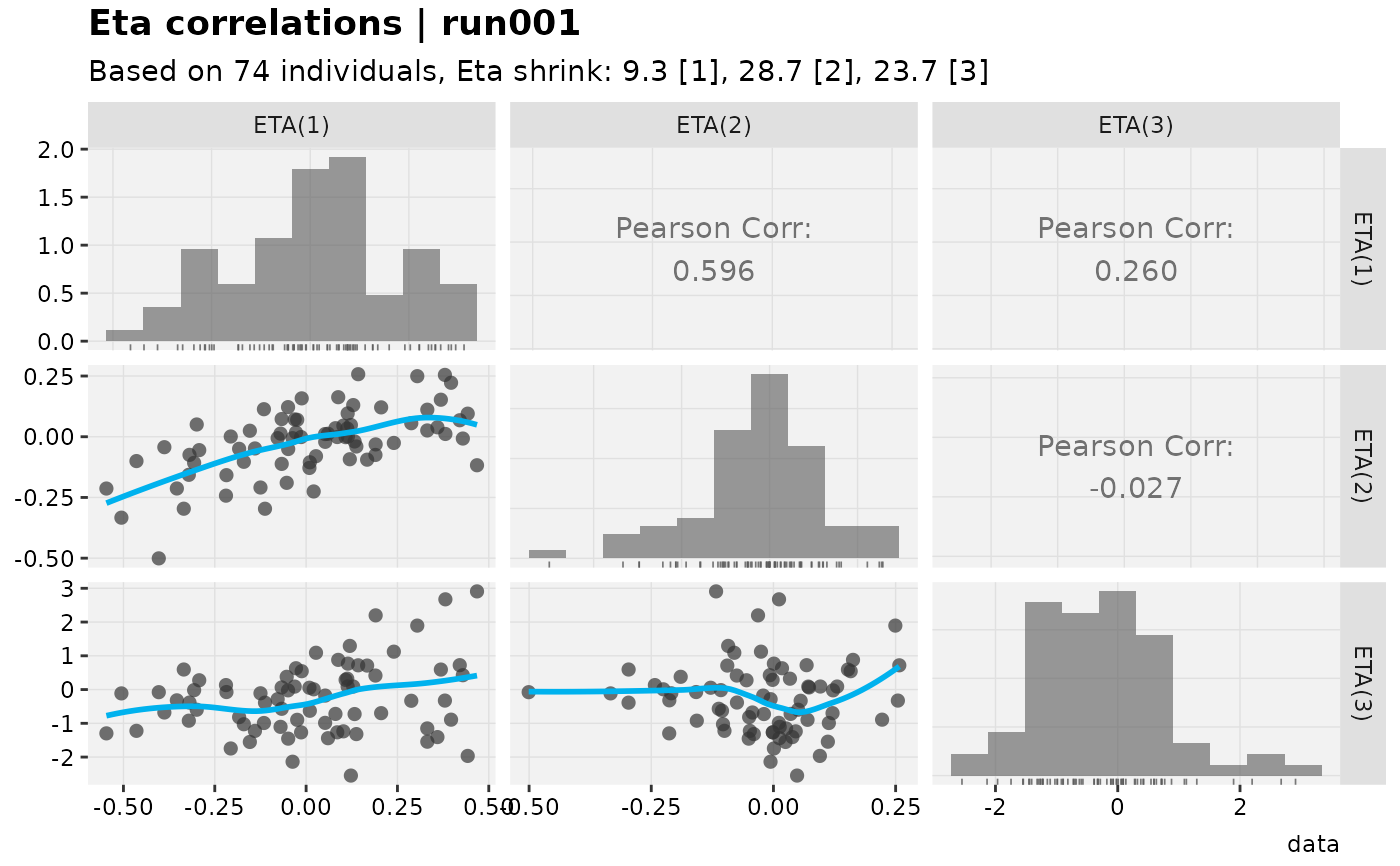
eta_grid(xpdb_x)
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
 cov_grid(xpdb_x)
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
cov_grid(xpdb_x)
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
 eta_vs_cov_grid(xpdb_x)
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
eta_vs_cov_grid(xpdb_x)
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
 # Labels and units are also supported
xpdb_x %>%
xpose::set_var_labels(AGE="Age", MED1 = "Digoxin") %>%
xpose::set_var_units(AGE="yrs") %>%
set_var_levels(SEX=lvl_sex(), MED1 = lvl_bin()) %>%
eta_vs_cov_grid()
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `out = purrr::map_if(...)`.
#> Caused by warning:
#> ! In $prob no.2 columns: MED1 not present in the data.
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
# Labels and units are also supported
xpdb_x %>%
xpose::set_var_labels(AGE="Age", MED1 = "Digoxin") %>%
xpose::set_var_units(AGE="yrs") %>%
set_var_levels(SEX=lvl_sex(), MED1 = lvl_bin()) %>%
eta_vs_cov_grid()
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `out = purrr::map_if(...)`.
#> Caused by warning:
#> ! In $prob no.2 columns: MED1 not present in the data.
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
 # }
# }
