nlmixr2 fits are fully supported in the current release,
including many convenience functions to ensure consumed
nlmixr2 results are fully compatible with most all
available features of xpose and
xpose.xtras.
Basics
While the xpose.xtras package already contains a few
nlmixr2 examples, let’s start fresh with a new fit. The
model below is from the
nlmixr2 documentation; to start using the resulting
xpose_data directly, refer to
?nlmixr2_warfarin.
pk.turnover.emax3 <- function() {
ini({
tktr <- log(1)
tka <- log(1)
tcl <- log(0.1)
tv <- log(10)
##
eta.ktr ~ 1
eta.ka ~ 1
eta.cl ~ 2
eta.v ~ 1
prop.err <- 0.1
pkadd.err <- 0.1
##
temax <- logit(0.8)
tec50 <- log(0.5)
tkout <- log(0.05)
te0 <- log(100)
##
eta.emax ~ .5
eta.ec50 ~ .5
eta.kout ~ .5
eta.e0 ~ .5
##
pdadd.err <- 10
})
model({
ktr <- exp(tktr + eta.ktr)
ka <- exp(tka + eta.ka)
cl <- exp(tcl + eta.cl)
v <- exp(tv + eta.v)
emax = expit(temax+eta.emax)
ec50 = exp(tec50 + eta.ec50)
kout = exp(tkout + eta.kout)
e0 = exp(te0 + eta.e0)
##
DCP = center/v
PD=1-emax*DCP/(ec50+DCP)
##
effect(0) = e0
kin = e0*kout
##
d/dt(depot) = -ktr * depot
d/dt(gut) = ktr * depot -ka * gut
d/dt(center) = ka * gut - cl / v * center
d/dt(effect) = kin*PD -kout*effect
##
cp = center / v
cp ~ prop(prop.err) + add(pkadd.err)
effect ~ add(pdadd.err) | pca
})
}
fit.TOS <- nlmixr2(pk.turnover.emax3, warfarin, "focei", control=list(print=0),
table=list(cwres=TRUE, npde=TRUE))
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> calculating covariance matrix
#> [====|====|====|====|====|====|====|====|====|====] 0:01:23
#> doneNow that we have a fit, we have a few options to consume the results
into xpose. We could use
xpose.nlmixr2::xpose_data_nlmixr2 and then convert to an
xp_xtras object from there.
xpose.nlmixr2::xpose_data_nlmixr2(fit.TOS) %>%
as_xp_xtras()
#>
#> ── ~ xp_xtras object
#> Model description: not implemented
#> fit.TOS overview:
#> - Software: nlmixr2 4.1.1
#> - Attached files (memory usage 553.8 Kb):
#> + obs tabs: $prob no.1: nlmixr2
#> + sim tabs: <none>
#> + output files: <none>
#> + special: <none>
#> + fit: <none>
#> - gg_theme: :: xpose theme_readable
#> - xp_theme: xp_xtra_theme new_x$xp_theme
#> - Options: dir = NULL, quiet = TRUE, manual_import = NULL, cvtype = exactIt is helpful though, for traceability and for some convenience
functions for the fit object to be attached to the
xpose_data object.
xpose.nlmixr2::xpose_data_nlmixr2(fit.TOS) %>%
as_xp_xtras() %>%
attach_nlmixr2(fit.TOS)
#>
#> ── ~ xp_xtras object
#> Model description: not implemented
#> fit.TOS overview:
#> - Software: nlmixr2 4.1.1
#> - Attached files (memory usage 725.7 Kb):
#> + obs tabs: $prob no.1: nlmixr2
#> + sim tabs: <none>
#> + output files: <none>
#> + special: <none>
#> + fit: attached as (this)$fit
#> - gg_theme: :: xpose theme_readable
#> - xp_theme: xp_xtra_theme new_x$xp_theme
#> - Options: dir = NULL, quiet = TRUE, manual_import = NULL, cvtype = exactOne such convenience function is to backfill some of the
"na" properties pulled from the fit results (at time of
writing, these are condition number and significant digits). It’s
acknowledged that these backfills are opinionated, hence it is
optional.
xpose.nlmixr2::xpose_data_nlmixr2(fit.TOS) %>%
as_xp_xtras() %>%
attach_nlmixr2(fit.TOS) %>%
backfill_nlmixr2_props() %>%
{print(get_prop(., "condn")); .} %>%
get_prop("nsig")
#> [1] "32318.0467755017"
#> [1] "3"A more useful application of the attached fit result is a method to
make get_prm() work. Note, xpose::prm_table()
does not work despite this new feature because it uses the original
get_prm() defined within xpose namespace.
Since prm_table() is a quick check convenience function to
print in place, get_prm() was considered a better function
to get working.
try({
xpose.nlmixr2::xpose_data_nlmixr2(fit.TOS) %>%
as_xp_xtras() %>% xpose::get_prm()
})
#> Error : No `files` slot could be found in this xpdb.
xpose.nlmixr2::xpose_data_nlmixr2(fit.TOS) %>%
as_xp_xtras() %>%
attach_nlmixr2(fit.TOS) %>%
get_prm() %>%
# Remove some columns for readability
dplyr::select(-c(fixed,diagonal,label))
#> # A tibble: 20 × 9
#> type name value se rse m n cv shk
#> <chr> <chr> <num:3> <num:3> <num:3> <int> <int> <num:3> <num:3>
#> 1 the tktr 0.104 2.20 21.1 1 NA NA NA
#> 2 the tka 0.302 2.18 7.23 2 NA NA NA
#> 3 the tcl -2.04 0.109 0.0536 3 NA NA NA
#> 4 the tv 2.06 0.0916 0.0444 4 NA NA NA
#> 5 the prop.err 0.148 NA NA 5 NA NA NA
#> 6 the pkadd.err 0.172 NA NA 6 NA NA NA
#> 7 the temax 4.75 6.20 1.30 7 NA NA NA
#> 8 the tec50 0.157 0.229 1.46 8 NA NA NA
#> 9 the tkout -2.93 0.128 0.0436 9 NA NA NA
#> 10 the te0 4.57 0.0399 0.00874 10 NA NA NA
#> 11 the pdadd.err 3.76 NA NA 11 NA NA NA
#> 12 ome eta.ktr 0.840 NA NA 1 1 101. 62.3
#> 13 ome eta.ka 0.944 NA NA 2 2 120. 60.6
#> 14 ome eta.cl 0.268 NA NA 3 3 27.3 -0.1
#> 15 ome eta.v 0.221 NA NA 4 4 22.4 10.3
#> 16 ome eta.emax 0.590 NA NA 5 5 64.5 95.1
#> 17 ome eta.ec50 0.453 NA NA 6 6 47.7 5.2
#> 18 ome eta.kout 0.153 NA NA 7 7 15.4 32.3
#> 19 ome eta.e0 0.103 NA NA 8 8 10.3 39.6
#> 20 sig sigma(1,1) 1 NA NA 1 1 NA 10.7To build on this, because nlmixr2 coerces users to use
mu-referencing and captures parameter associations automatically,
another useful backfill includes mapping these parameter associations
with add_prm_association(), by way of
nlmixr2_prm_associations(). In this model,
temax is logit transformed, so to keep assumptions valid in
the CV% calculation it needs to be back-transformed (as indicated in a
cli message).
xpose.nlmixr2::xpose_data_nlmixr2(fit.TOS) %>%
as_xp_xtras() %>%
attach_nlmixr2(fit.TOS) %>%
nlmixr2_prm_associations() %>%
mutate_prm(temax ~ plogis) %>%
get_prm() %>%
# Remove some columns for readability
dplyr::select(-c(fixed,diagonal,label))
#> # A tibble: 20 × 9
#> type name value se rse m n cv shk
#> <chr> <chr> <num:3> <num:3> <num:3> <int> <int> <num:3> <num:3>
#> 1 the tktr 0.104 2.20 21.1 1 NA NA NA
#> 2 the tka 0.302 2.18 7.23 2 NA NA NA
#> 3 the tcl -2.04 0.109 0.0536 3 NA NA NA
#> 4 the tv 2.06 0.0916 0.0444 4 NA NA NA
#> 5 the prop.err 0.148 NA NA 5 NA NA NA
#> 6 the pkadd.err 0.172 NA NA 6 NA NA NA
#> 7 the temax 0.991 0.362 0.365 7 NA NA NA
#> 8 the tec50 0.157 0.229 1.46 8 NA NA NA
#> 9 the tkout -2.93 0.128 0.0436 9 NA NA NA
#> 10 the te0 4.57 0.0399 0.00874 10 NA NA NA
#> 11 the pdadd.err 3.76 NA NA 11 NA NA NA
#> 12 ome eta.ktr 0.840 NA NA 1 1 101. 62.3
#> 13 ome eta.ka 0.944 NA NA 2 2 120. 60.6
#> 14 ome eta.cl 0.268 NA NA 3 3 27.3 -0.1
#> 15 ome eta.v 0.221 NA NA 4 4 22.4 10.3
#> 16 ome eta.emax 0.590 NA NA 5 5 0.647 95.1
#> 17 ome eta.ec50 0.453 NA NA 6 6 47.7 5.2
#> 18 ome eta.kout 0.153 NA NA 7 7 15.4 32.3
#> 19 ome eta.e0 0.103 NA NA 8 8 10.3 39.6
#> 20 sig sigma(1,1) 1 NA NA 1 1 NA 10.7
#> # Parameter table includes the following associations: tktr~log(eta.ktr),
#> tka~log(eta.ka), tcl~log(eta.cl), tv~log(eta.v), temax~logit(eta.emax),
#> tec50~log(eta.ec50), tkout~log(eta.kout), and te0~log(eta.e0)That’s a lot of boilerplate to pipe through, so a convenience
function has been developed to cover all of that. Note the parameter
associations are mapped automatically, but we could skip that step if we
weren’t planning to use get_prm() at all by setting
.skip_assoc=TRUE.
nlmixr2_warfarin <- nlmixr2_as_xtra(fit.TOS)General Usage
We covered most of the usage unique to this package in the Basics
section, but this section is to illustrate that common functions will
continue to work. At time of writing, some missing properties are
present and should be added with a PR to
xpose.nlmixr2 (these are not backfill candidates since they
don’t exist in the summary element, violating how
set_prop() was designed to work).
list_vars(nlmixr2_warfarin)
#> List of available variables for problem no. 1
#> - Subject identifier (id) : ID
#> - Dependent variable (dv) : DV
#> - Independent variable (idv) : TIME
#> - DV identifier (dvid) : DVID [0]
#> - Dose amount (amt) : AMT
#> - Event identifier (evid) : EVID
#> - Model typical predictions (pred) : CPRED
#> - Model individual predictions (ipred) : IPRED
#> - Model parameter (param) : KA, CL, V
#> - Eta (eta) : eta.ktr, eta.ka, eta.cl, eta.v, eta.emax, eta.ec50, eta.kout, eta.e0
#> - Residuals (res) : NPDE, RES, WRES, IRES, IWRES, CRES, CWRES
#> - Categorical covariates (catcov) : SEX [0]
#> - Continuous covariates (contcov) : WT, AGE
#> - Not attributed (na) : NLMIXRLLIKOBS, CMT, EPRED, ERES, NPD, PDE, PD, PRED, ETA.KTR, ETA.KA, ETA.CL, ETA.V, ETA.EMAX, ETA.EC50, ETA.KOUT, ETA.E0, DEPOT, GUT, CENTER, EFFECT, KTR, EMAX, EC50, KOUT, E0, DCP, PD.1, KIN, TAD, DOSENUM
dv_vs_ipred(nlmixr2_warfarin, facet="DVID")
#> `geom_smooth()` using formula = 'y ~ x'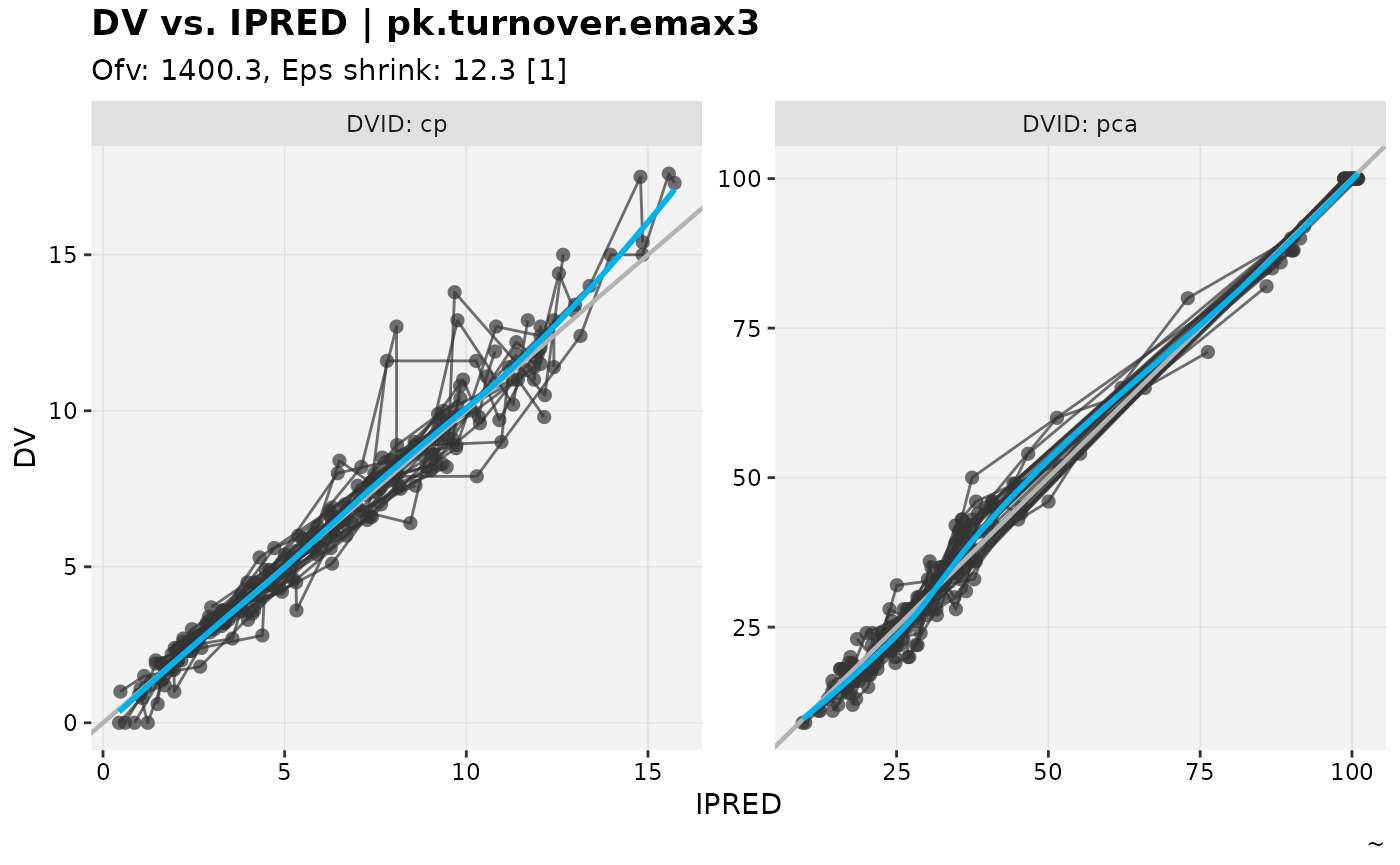
dv_vs_pred(nlmixr2_warfarin, facet="DVID")
#> `geom_smooth()` using formula = 'y ~ x'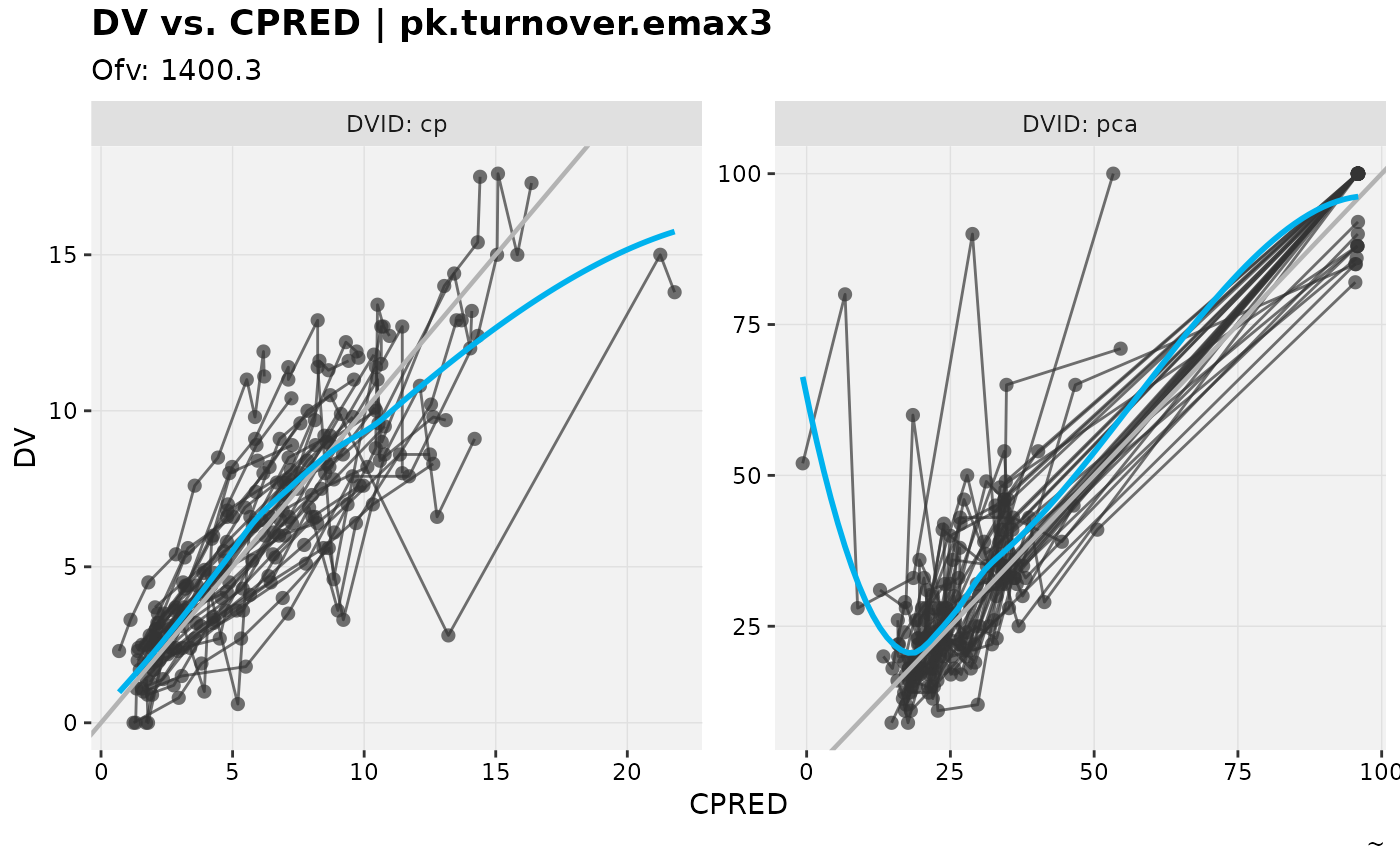
eta_vs_catcov(nlmixr2_warfarin, etavar = eta.cl)
#> Warning: nind is not part of the available keywords. Check ?template_titles for
#> a full list.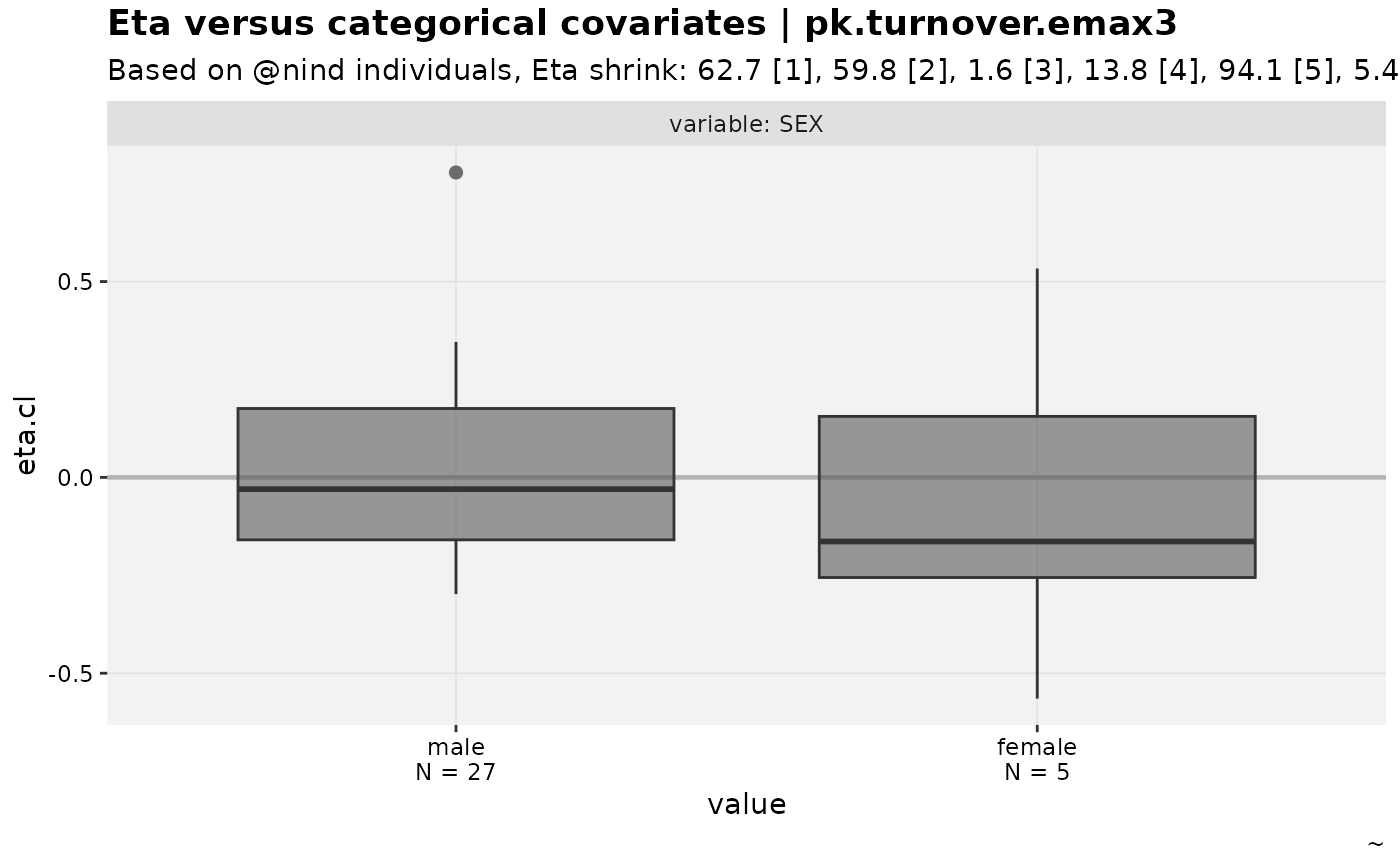
eta_vs_contcov(nlmixr2_warfarin, etavar = eta.cl)
#> Warning: nind is not part of the available keywords. Check ?template_titles for
#> a full list.
#> `geom_smooth()` using formula = 'y ~ x'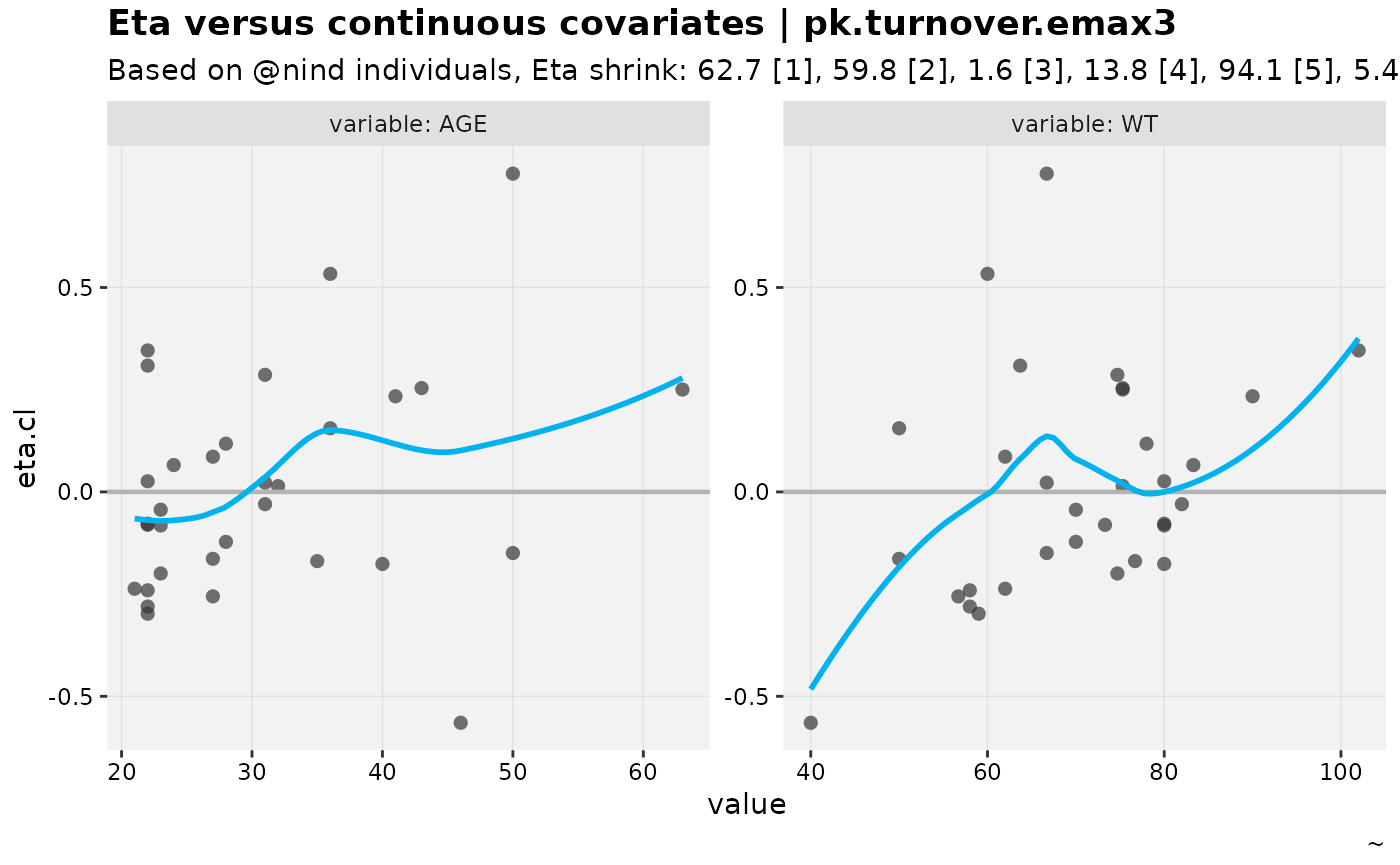
eta_vs_cov_grid(nlmixr2_warfarin, etavar = c(eta.cl,eta.v,eta.ka), quiet=TRUE)
#> Warning: nind is not part of the available keywords. Check ?template_titles for
#> a full list.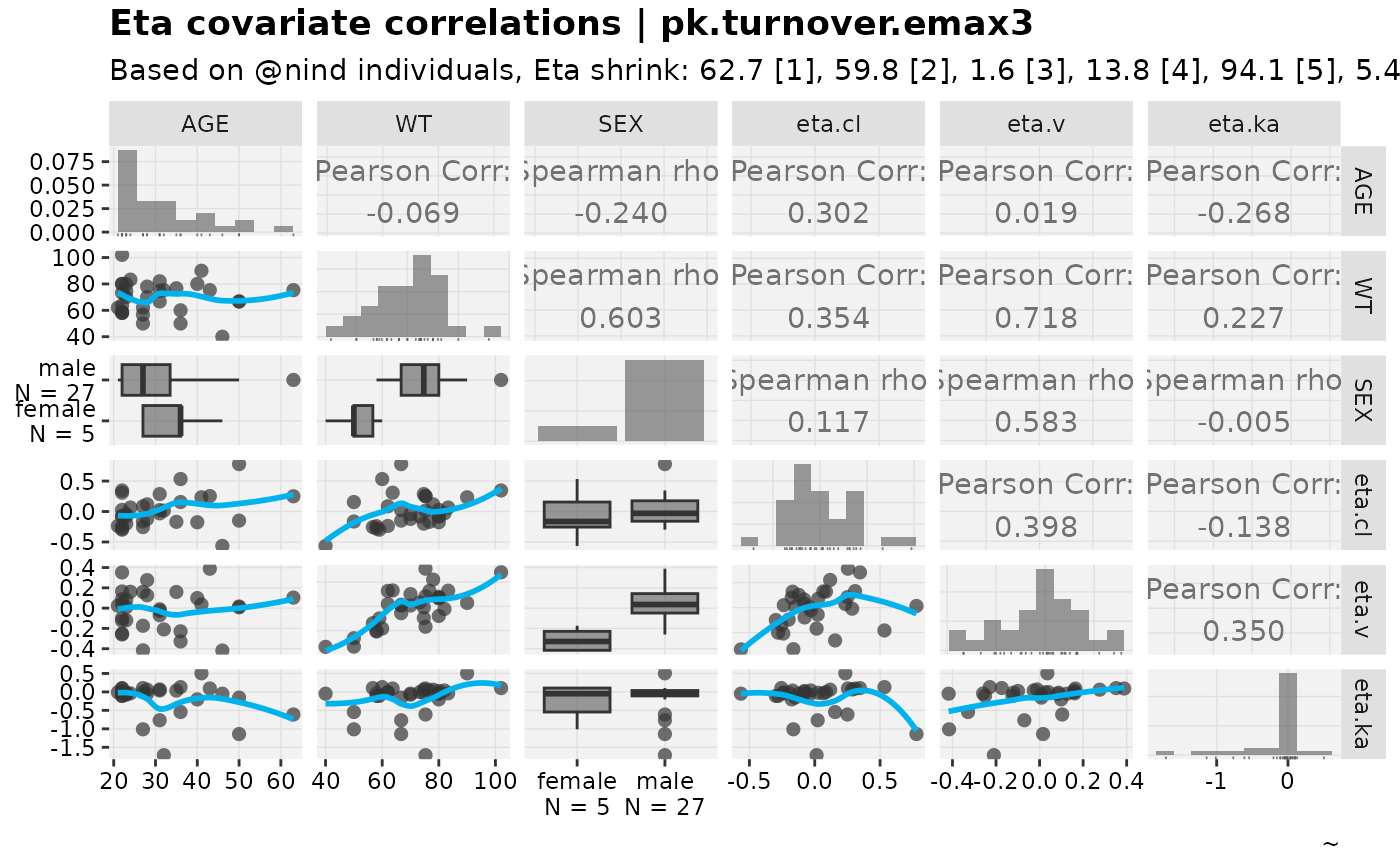
eta_vs_cov_grid(nlmixr2_warfarin, etavar = c(eta.kout,eta.e0,eta.emax), quiet=TRUE)
#> Warning: nind is not part of the available keywords. Check ?template_titles for
#> a full list.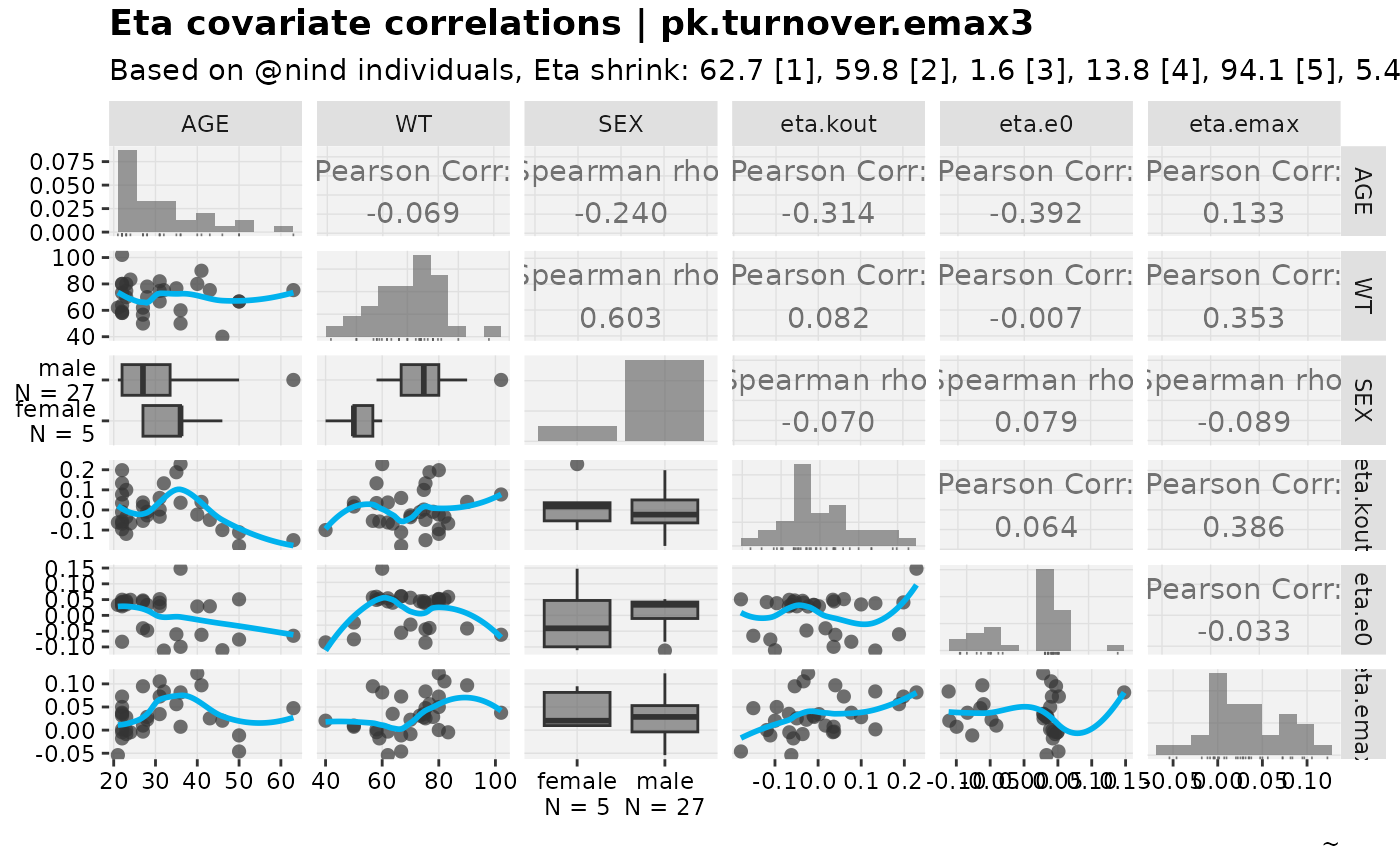
Sets
A simple change to the warfarin PK model is to use the same
parameters for ka and ktr. In the PD model, we
could also fix emax to 1 (and drop the
interindividual variability). An analyst would explore these separately
and then maybe combine them, as we have done below.
fit.TOS.kaktr <- fit.TOS %>%
model({
ka <- ktr
}) %>%
nlmixr2(warfarin, "focei",
control = list(print = 0),
table = list(cwres = TRUE, npde = TRUE)
)
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> calculating covariance matrix
#> [====|====|====|====|====|====|====|====|====|====] 0:00:51
#> done
fit.TOS.emax1 <- fit.TOS %>%
model({
emax = 1
}) %>%
nlmixr2(warfarin, "focei",
control = list(print = 0),
table = list(cwres = TRUE, npde = TRUE)
)
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> calculating covariance matrix
#> [====|====|====|====|====|====|====|====|====|====] 0:00:54
#> done
fit.TOS.simple <- fit.TOS %>%
model({
ka <- ktr
emax = 1
}) %>%
nlmixr2(warfarin, "focei",
control = list(print = 0),
table = list(cwres = TRUE, npde = TRUE)
)
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#>
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> [====|====|====|====|====|====|====|====|====|====] 0:00:00
#> calculating covariance matrix
#> [====|====|====|====|====|====|====|====|====|====] 0:00:33
#> doneThese explorations make up a set which can be examined with an
xpose_set:
# Since these models were created with piping, need to update their labels
warf_ka <- nlmixr2_as_xtra(fit.TOS.kaktr) %>%
set_prop(run="fit.TOS.kaktr") %>%
# Just to remove github action path
set_option(
dir = paste0("~")
) %>%
set_prop(
dir = paste0("~")
)
warf_emax <- nlmixr2_as_xtra(fit.TOS.emax1) %>%
set_prop(run="fit.TOS.emax1") %>%
set_option(
dir = paste0("~")
) %>%
set_prop(
dir = paste0("~")
)
warf_simple <- nlmixr2_as_xtra(fit.TOS.simple) %>%
set_prop(run="fit.TOS.simple") %>%
set_option(
dir = paste0("~")
) %>%
set_prop(
dir = paste0("~")
)
warfarin_set <- xpose_set(
nlmixr2_warfarin, warf_ka, warf_emax, warf_simple,
.relationships = c(
warf_ka~nlmixr2_warfarin,
warf_emax~nlmixr2_warfarin,
warf_simple ~ warf_ka + warf_emax
)
) %>%
# Add iOFVs
focus_qapply(backfill_iofv)
warfarin_set
#>
#> ── xpose_set object ────────────────────────────────────────────────────────────
#> • Number of models: 4
#> • Model labels: nlmixr2_warfarin, warf_ka, warf_emax, and warf_simple
#> • Number of relationships: 4
#> • Focused xpdb objects: none
#> • Exposed properties: none
#> • Base model: none
warfarin_set$warf_simple
#>
#> ── Part of an xpose_set, with label: warf_simple ───────────────────────────────
#> • Parent(s): warf_ka and warf_emax
#> • In focus?: no
#> • Base model?: no
#>
#> ── xpdb object (accessible with {xpose_set}$warf_simple$xpdb):
#>
#> ── ~ xp_xtras object
#> Model description: not implemented
#> fit.TOS.simple overview:
#> - Software: nlmixr2 4.1.1
#> - Attached files (memory usage 693.4 Kb):
#> + obs tabs: $prob no.1 (modified): na, nlmixr2
#> + sim tabs: <none>
#> + output files: <none>
#> + special: <none>
#> + fit: attached as (this)$fit
#> - gg_theme: :: xpose theme_readable
#> - xp_theme: xp_xtra_theme new_x$xp_theme
#> - Options: dir = ~, quiet = TRUE, manual_import = NULL, cvtype = exact
warfarin_set %>%
expose_property(ofv) %>%
expose_property(condn) %>%
reshape_set() %>%
# Remove some columns for readability
dplyr::select(-c(parent,base,focus))
#> # A tibble: 4 × 4
#> xpdb label ..ofv ..condn
#> <named list> <chr> <dbl> <dbl>
#> 1 <xp_xtras> nlmixr2_warfarin 1387 7497.
#> 2 <xp_xtras> warf_ka 1338. 21266.
#> 3 <xp_xtras> warf_emax 1351. 464.
#> 4 <xp_xtras> warf_simple 1338. 269.
warfarin_set %>%
dofv_vs_id(nlmixr2_warfarin, warf_simple, .inorder = TRUE, df=1)
#> Warning: nind is not part of the available keywords. Check ?template_titles for
#> a full list.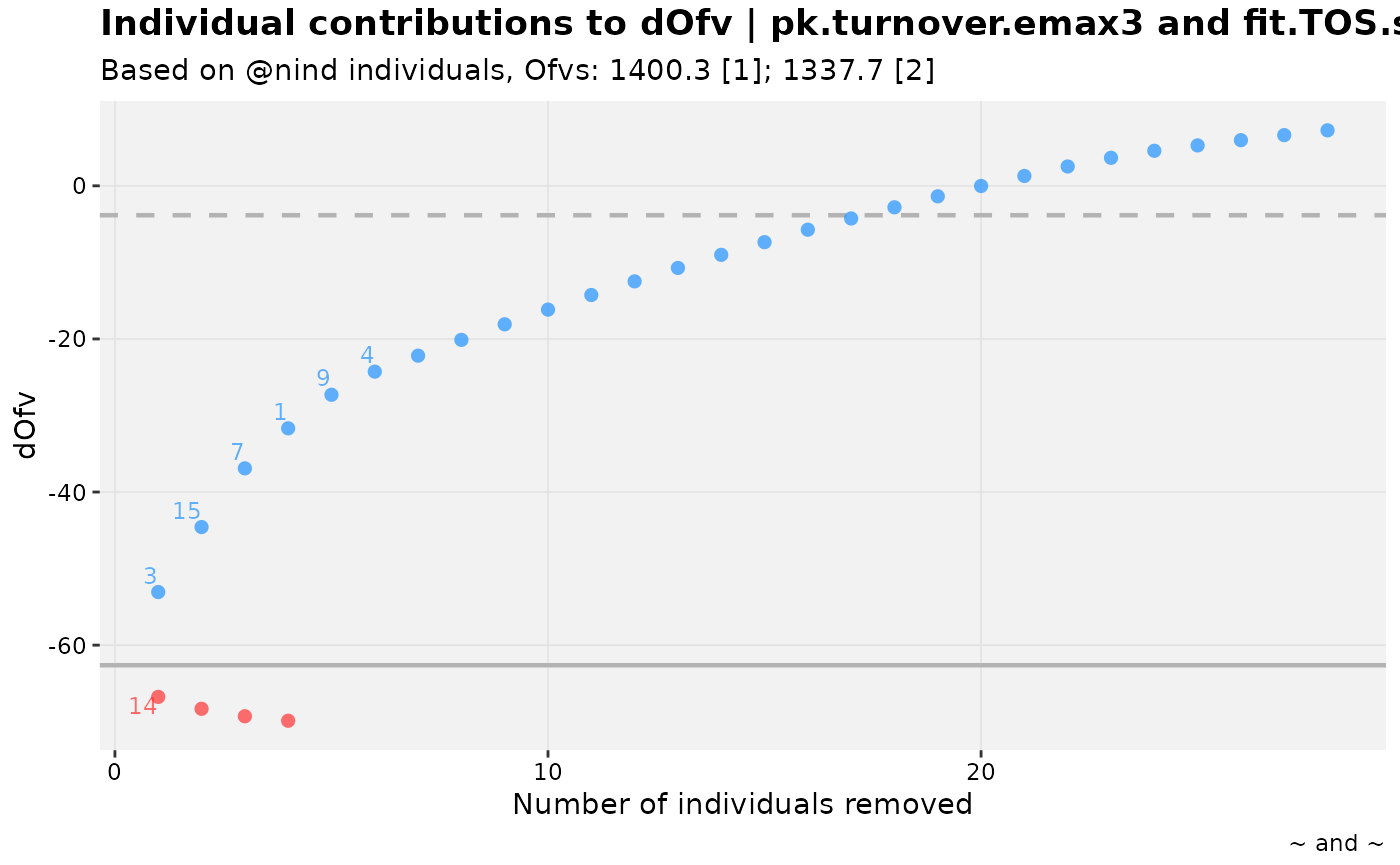
warfarin_set %>%
# This comparison should be more interesting
focus_qapply(set_var_types, param=c(KA,EC50), na=c(CL, V)) %>%
prm_waterfall(nlmixr2_warfarin, warf_simple)
#> Warning: nind is not part of the available keywords. Check ?template_titles for
#> a full list.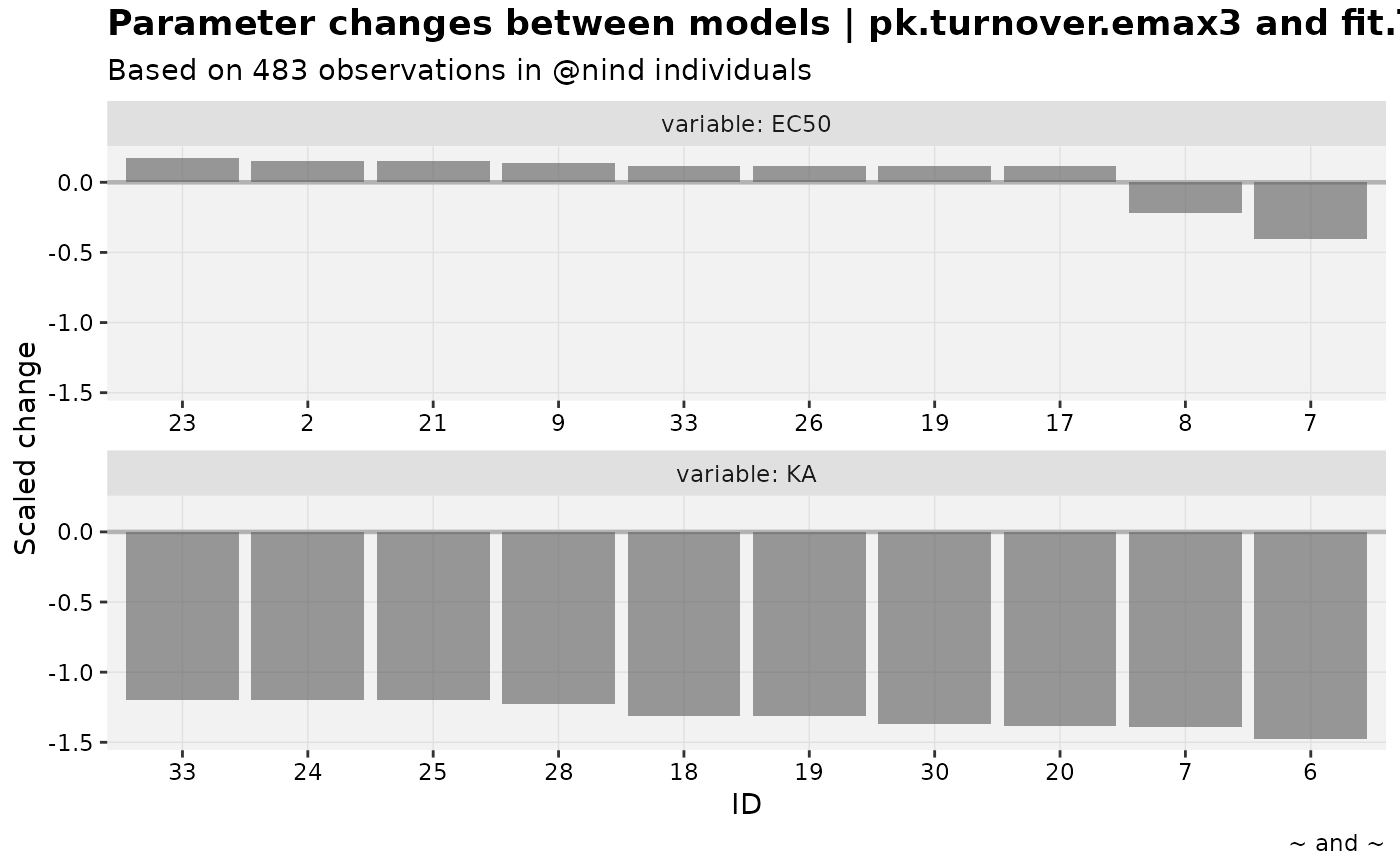
warfarin_set %>%
dv_vs_ipred_modavg(warf_emax, warf_ka)
#> `geom_smooth()` using formula = 'y ~ x'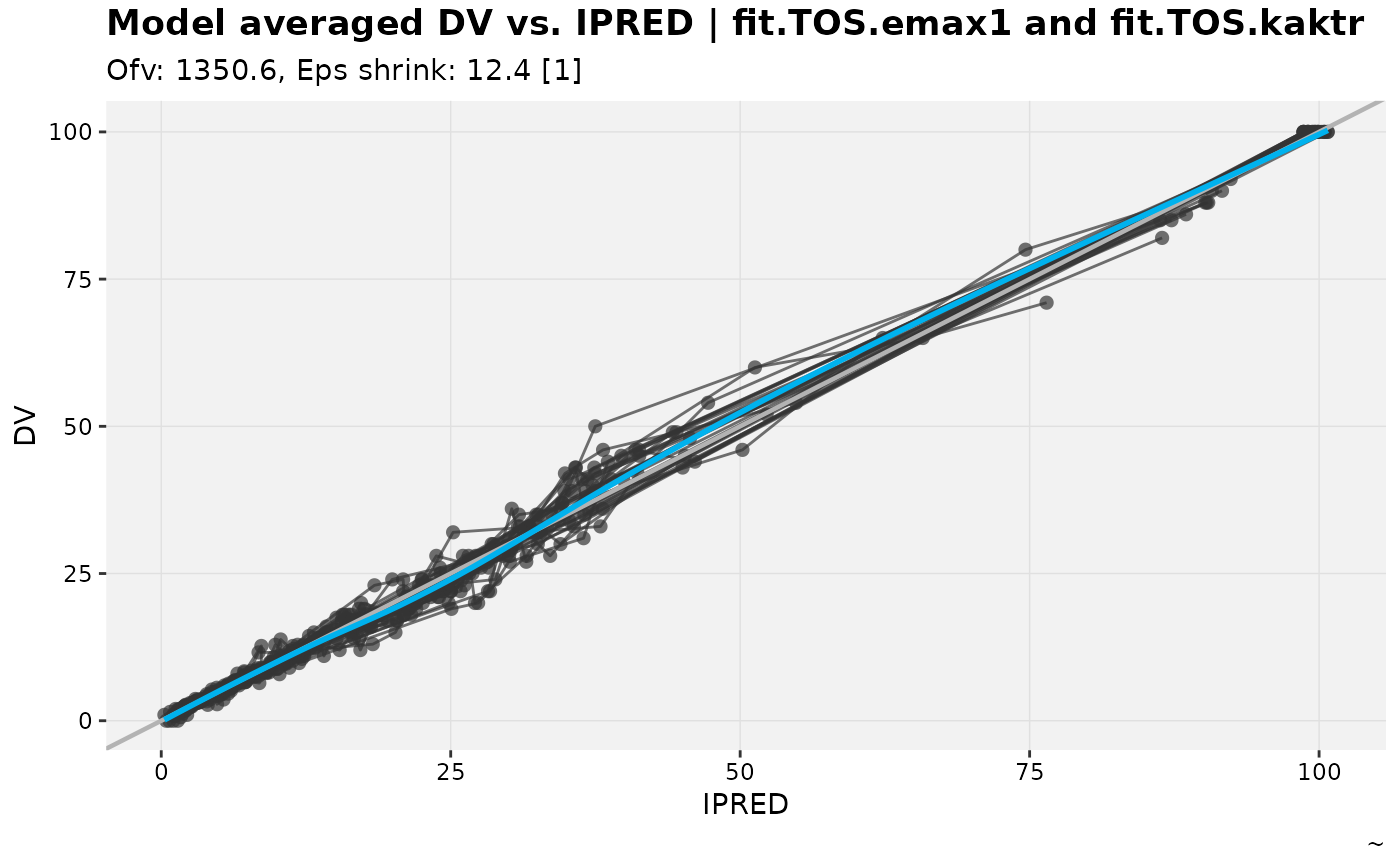
Derived parameter explorations
The suggestion to have nlmixr2 installed enables
rxode2 utility functions to be leveraged for diagnostics
(and probably many other unrealized benefits, possibly using
nonmem2rx). One of those features is exploring the derived
parameters for a model.
Now any model (it does not have to be fit with nlmixr2)
can have derived parameters calculated and checked for signs of
misspecification. The derive_prm() family of functions can
be used to generate a table of derived parameters or to set derived
parameters as param type variables.
nlmixr2_m3 %>%
backfill_derived() %>%
list_vars()
#> List of available variables for problem no. 1
#> - Subject identifier (id) : ID
#> - Dependent variable (dv) : DV
#> - Independent variable (idv) : TIME
#> - Dose amount (amt) : AMT
#> - Event identifier (evid) : EVID
#> - Model typical predictions (pred) : CPRED
#> - Model individual predictions (ipred) : IPRED
#> - Model parameter (param) : KA, CL, V, VC, KEL, VSS, T12ALPHA, ALPHA, A, FRACA
#> - Eta (eta) : eta.ka, eta.cl, eta.v
#> - Residuals (res) : RES, WRES, IRES, IWRES, CRES, CWRES
#> - Continuous covariates (contcov) : WT
#> - Not attributed (na) : CMT, CENS, LLOQ, NLMIXRLLIKOBS, PRED, LOWERLIM, UPPERLIM, ETA.KA, ETA.CL, ETA.V, DEPOT, CENT, BLQLIKE, TAD, DOSENUM
derive_prm(nlmixr2_m3) %>%
dplyr::select(ID,KA,CL,VSS:(dplyr::last_col())) %>%
head()
#> ID KA CL VSS T12ALPHA ALPHA A FRACA
#> 1 1 1.7262871 1.725758 28.98448 11.641557 0.05954076 0.03450122 1
#> 2 10 0.7714723 1.880091 26.90314 9.918579 0.06988372 0.03717039 1
#> 3 11 3.2255861 3.788283 35.71719 6.535222 0.10606329 0.02799772 1
#> 4 12 0.9557512 2.423387 26.15672 7.481452 0.09264875 0.03823110 1
#> 5 2 1.8922247 3.262844 31.51880 6.695745 0.10352055 0.03172709 1
#> 6 3 2.2187556 2.933599 32.93694 7.782299 0.08906715 0.03036105 1
# If param has no vars, .prm should be set
pheno_base %>%
backfill_derived(
.prm = c(CL,V)
) %>%
list_vars()
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> List of available variables for problem no. 1
#> - Subject identifier (id) : ID
#> - Dependent variable (dv) : DV
#> - Independent variable (idv) : TIME
#> - Dose amount (amt) : AMT
#> - Event identifier (evid) : EVID
#> - Missing dependent variable (mdv) : MDV
#> - Model typical predictions (pred) : PRED
#> - Model individual predictions (ipred) : IPRED
#> - Model parameter (param) : CL, V, VC, KEL, VSS, T12ALPHA, ALPHA, A, FRACA
#> - Eta (eta) : ETA1, ETA2
#> - Residuals (res) : IWRES, CWRES, NPDE, RES, WRES
#> - Categorical covariates (catcov) : APGR ('Apgar score') [10]
#> - Continuous covariates (contcov) : WT ('Weight', kg)
#> - Not attributed (na) : IRES, CRESThese derived parameters can be fed into
diagnose_constants() to perform some quick checks for
common issues. In this case, we have both VC (derived) and
V (fitted) representing the same quantity, so to avoid
catching both in the volume check (which per the documentation should
only be one volume) the vol_pattern has been updated.
nlmixr2_m3 %>%
backfill_derived() %>%
diagnose_constants(vol_pattern = "^V$")
#> ℹ Checking for absorption flip-flop (first-order absorption slower than derived rate constants)...
#> ✔ No parameter sets are suggestive of flip-flop.
#> ℹ Checking for negative microconstants or volume...
#> ✔ No parameter sets have negative microconstants or volumes.
nlmixr2_m3 %>%
backfill_derived() %>%
diagnose_constants(
vol_pattern = "^V$",
df_units = list(KA = "1/hr", ALPHA = "1/hr"),
checks = list(neg_microvol = FALSE)
)
#> ℹ Checking for absorption flip-flop (first-order absorption slower than derived rate constants)...
#> ✔ No parameter sets are suggestive of flip-flop.
#> ℹ Checking that compared units match...
#> ✔ All relevant units seem to match.
# Using df form
derive_prm(nlmixr2_m3) %>%
diagnose_constants(df = ., vol_pattern = "^V$")
#> ℹ Checking for absorption flip-flop (first-order absorption slower than derived rate constants)...
#> ✔ No parameter sets are suggestive of flip-flop.
#> ℹ Checking for negative microconstants or volume...
#> ✔ No parameter sets have negative microconstants or volumes.Session info
sessionInfo()
#> R version 4.5.2 (2025-10-31)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 24.04.3 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] xpose.xtras_0.1.2 xpose.nlmixr2_0.4.1 xpose_0.4.22
#> [4] ggplot2_4.0.1 rxode2_4.1.1 nlmixr2plot_3.0.3
#> [7] nlmixr2extra_3.0.2 nlmixr2est_4.1.1 nlmixr2data_2.0.9
#> [10] lotri_1.0.2 nlmixr2_4.0.1
#>
#> loaded via a namespace (and not attached):
#> [1] tidyselect_1.2.1 dplyr_1.1.4 farver_2.1.2
#> [4] S7_0.2.1 fastmap_1.2.0 lazyeval_0.2.2
#> [7] GGally_2.4.0 tweenr_2.0.3 rex_1.2.1
#> [10] stringfish_0.17.0 digest_0.6.39 lifecycle_1.0.4
#> [13] magrittr_2.0.4 dparser_1.3.1-13 compiler_4.5.2
#> [16] rlang_1.1.6 sass_0.4.10 tools_4.5.2
#> [19] utf8_1.2.6 yaml_2.3.10 data.table_1.17.8
#> [22] symengine_0.2.10 knitr_1.50 lbfgsb3c_2024-3.5
#> [25] labeling_0.4.3 htmlwidgets_1.6.4 pmxcv_0.0.2
#> [28] RColorBrewer_1.1-3 withr_3.0.2 purrr_1.2.0
#> [31] sys_3.4.3 desc_1.4.3 grid_4.5.2
#> [34] polyclip_1.10-7 scales_1.4.0 MASS_7.3-65
#> [37] cli_3.6.5 rmarkdown_2.30 crayon_1.5.3
#> [40] ragg_1.5.0 generics_0.1.4 RcppParallel_5.1.11-1
#> [43] rstudioapi_0.17.1 tzdb_0.5.0 cachem_1.1.0
#> [46] ggforce_0.5.0 RApiSerialize_0.1.4 stringr_1.6.0
#> [49] splines_4.5.2 vctrs_0.6.5 Matrix_1.7-4
#> [52] jsonlite_2.0.0 PreciseSums_0.7 hms_1.1.4
#> [55] systemfonts_1.3.1 tidyr_1.3.1 jquerylib_0.1.4
#> [58] rxode2ll_2.0.13 glue_1.8.0 pkgdown_2.2.0
#> [61] ggstats_0.11.0 codetools_0.2-20 stringi_1.8.7
#> [64] gtable_0.3.6 tibble_3.3.0 pillar_1.11.1
#> [67] htmltools_0.5.8.1 R6_2.6.1 textshaping_1.0.4
#> [70] conflicted_1.2.0 evaluate_1.0.5 lattice_0.22-7
#> [73] readr_2.1.6 backports_1.5.0 vpc_1.2.2
#> [76] qs_0.27.3 memoise_2.0.1 n1qn1_6.0.1-12
#> [79] bslib_0.9.0 Rcpp_1.1.0 nlme_3.1-168
#> [82] checkmate_2.3.3 mgcv_1.9-3 xfun_0.54
#> [85] fs_1.6.6 forcats_1.0.1 pkgconfig_2.0.3