Usage
dv_vs_ipred_modavg(
xpdb_s,
...,
.lineage = FALSE,
algorithm = c("maa", "msa"),
weight_type = c("individual", "population"),
auto_backfill = FALSE,
weight_basis = c("ofv", "aic", "res"),
res_col = "RES",
quiet
)
dv_vs_pred_modavg(
xpdb_s,
...,
.lineage = FALSE,
algorithm = c("maa", "msa"),
weight_type = c("individual", "population"),
auto_backfill = FALSE,
weight_basis = c("ofv", "aic", "res"),
res_col = "RES",
quiet
)
ipred_vs_idv_modavg(
xpdb_s,
...,
.lineage = FALSE,
algorithm = c("maa", "msa"),
weight_type = c("individual", "population"),
auto_backfill = FALSE,
weight_basis = c("ofv", "aic", "res"),
res_col = "RES",
quiet
)
pred_vs_idv_modavg(
xpdb_s,
...,
.lineage = FALSE,
algorithm = c("maa", "msa"),
weight_type = c("individual", "population"),
auto_backfill = FALSE,
weight_basis = c("ofv", "aic", "res"),
res_col = "RES",
quiet
)
plotfun_modavg(
xpdb_s,
...,
.lineage = FALSE,
avg_cols = NULL,
avg_by_type = NULL,
algorithm = c("maa", "msa"),
weight_type = c("individual", "population"),
auto_backfill = FALSE,
weight_basis = c("ofv", "aic", "res"),
res_col = "RES",
.fun = NULL,
.funargs = list(),
quiet
)Arguments
- xpdb_s
<
xpose_set> object- ...
<
tidyselect> of models in set. If empty, all models are used in order of their position in the set. May also use a formula, which will just be processed withall.vars().- .lineage
<
logical> where ifTRUE,...is processed- algorithm
<
character> Model selection or model averaging- weight_type
<
character> Individual-level averaging or by full dataset.- auto_backfill
<
logical> If true, <backfill_iofv> is automatically applied.- weight_basis
<
character> Weigh by OFV (default), AIC or residual.- res_col
<
character> Column to weight by if"res"weight basis.- quiet
<
logical> Minimize extra output.- avg_cols
<
tidyselect> columns in data to average- avg_by_type
<
character> Mainly for use in wrapper functions. Column type to average, but resulting column names must be valid foravg_cols(ie, same across all objects in the set).avg_colswill be overwritten.- .fun
<
function> For slightly more convenient piping of model-averagedxpose_datainto a plotting function.- .funargs
<
list> Extra args to pass to function. If passingtidyselectarguments, be mindful of where quosures might be needed. See Examples.
Examples
# \donttest{
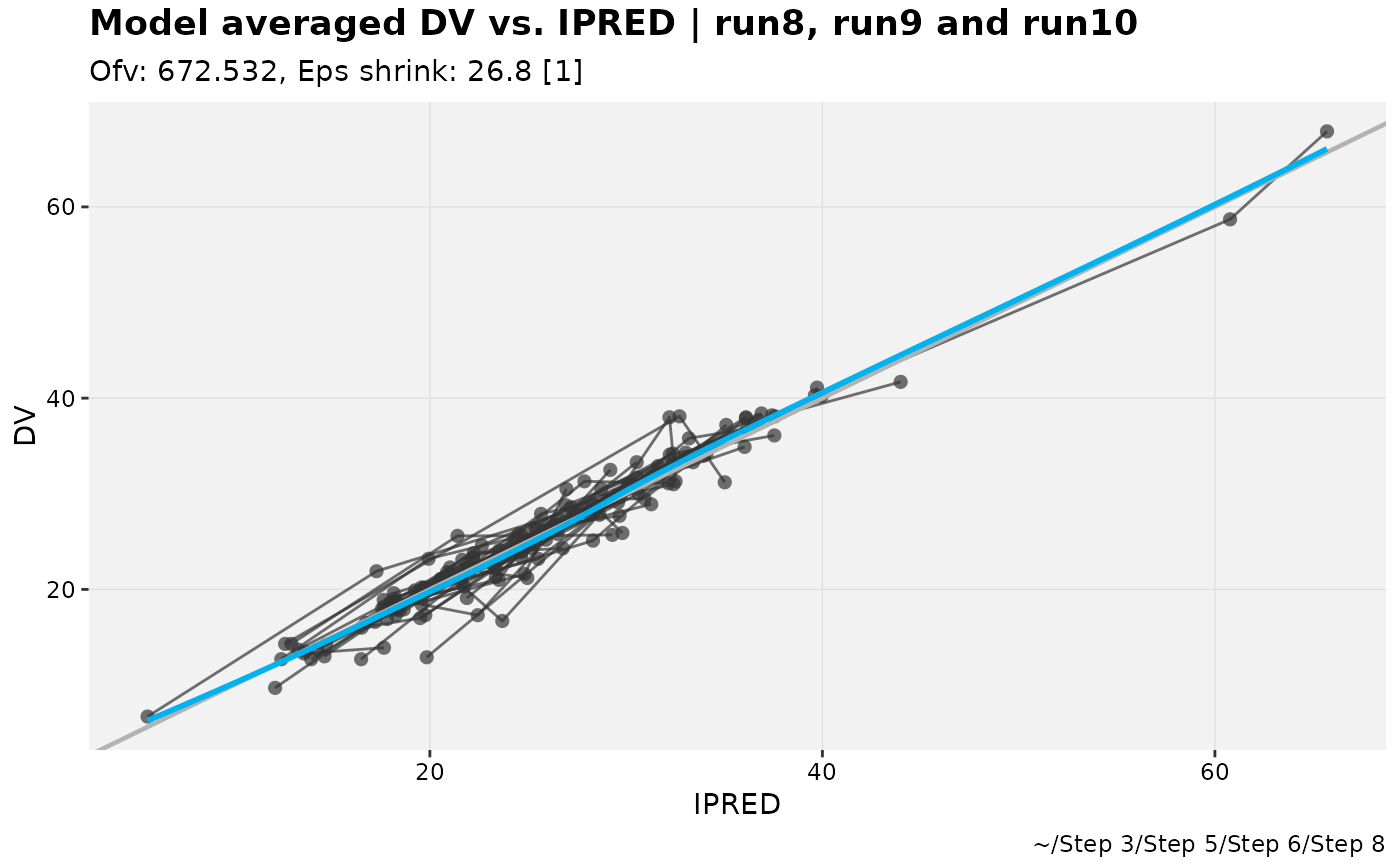
pheno_set %>%
dv_vs_ipred_modavg(run8,run9,run10, auto_backfill = TRUE)
#> Using data from $prob no.1
#> Filtering data by EVID == 0
#> `geom_smooth()` using formula = 'y ~ x'
 pheno_set %>%
dv_vs_pred_modavg(run8,run9,run10, auto_backfill = TRUE)
#> Using data from $prob no.1
#> Filtering data by EVID == 0
#> `geom_smooth()` using formula = 'y ~ x'
pheno_set %>%
dv_vs_pred_modavg(run8,run9,run10, auto_backfill = TRUE)
#> Using data from $prob no.1
#> Filtering data by EVID == 0
#> `geom_smooth()` using formula = 'y ~ x'
 pheno_set %>%
ipred_vs_idv_modavg(run8,run9,run10, auto_backfill = TRUE)
#> Using data from $prob no.1
#> Filtering data by EVID == 0
#> `geom_smooth()` using formula = 'y ~ x'
pheno_set %>%
ipred_vs_idv_modavg(run8,run9,run10, auto_backfill = TRUE)
#> Using data from $prob no.1
#> Filtering data by EVID == 0
#> `geom_smooth()` using formula = 'y ~ x'
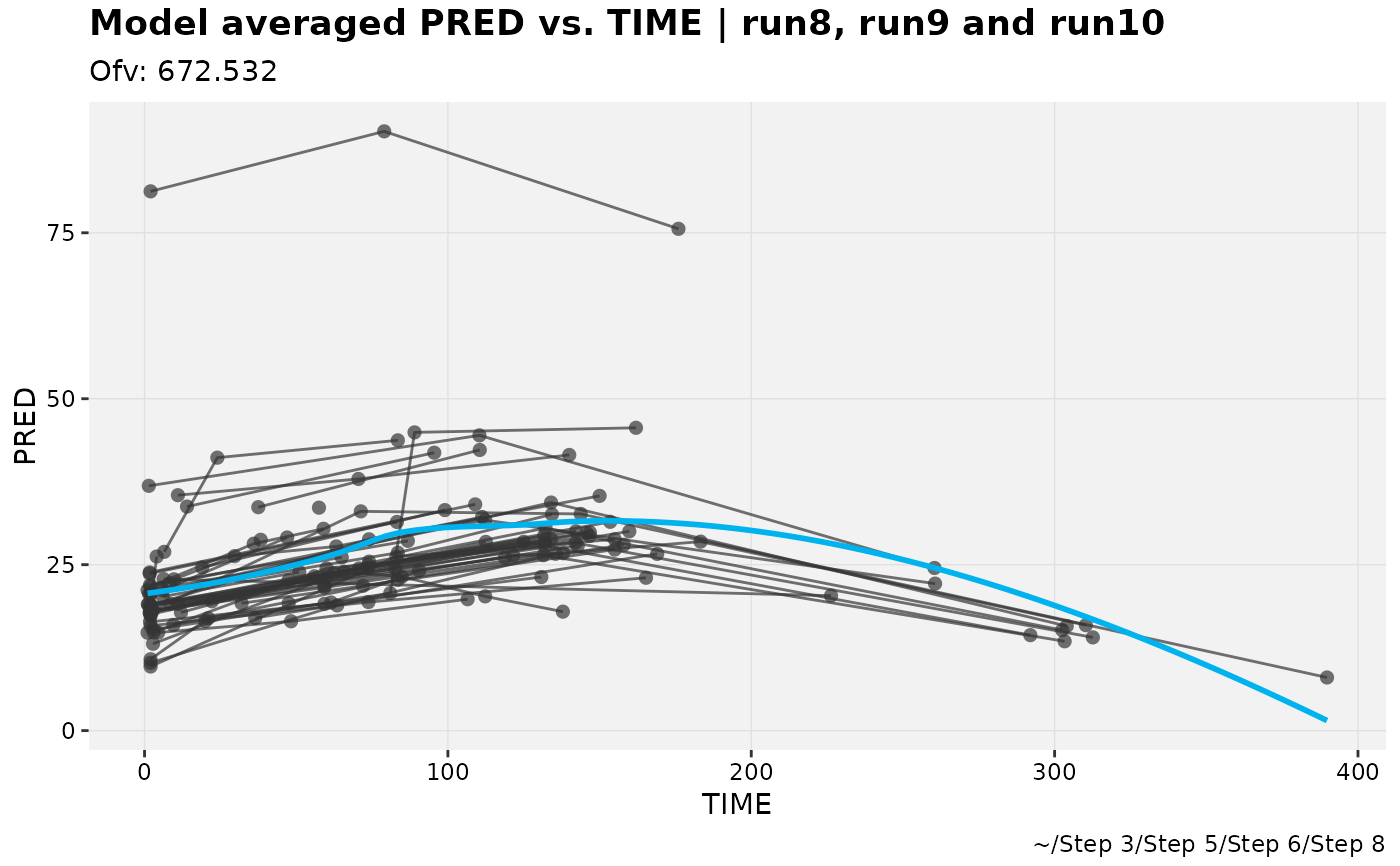
 pheno_set %>%
pred_vs_idv_modavg(run8,run9,run10, auto_backfill = TRUE)
#> Using data from $prob no.1
#> Filtering data by EVID == 0
#> `geom_smooth()` using formula = 'y ~ x'
pheno_set %>%
pred_vs_idv_modavg(run8,run9,run10, auto_backfill = TRUE)
#> Using data from $prob no.1
#> Filtering data by EVID == 0
#> `geom_smooth()` using formula = 'y ~ x'
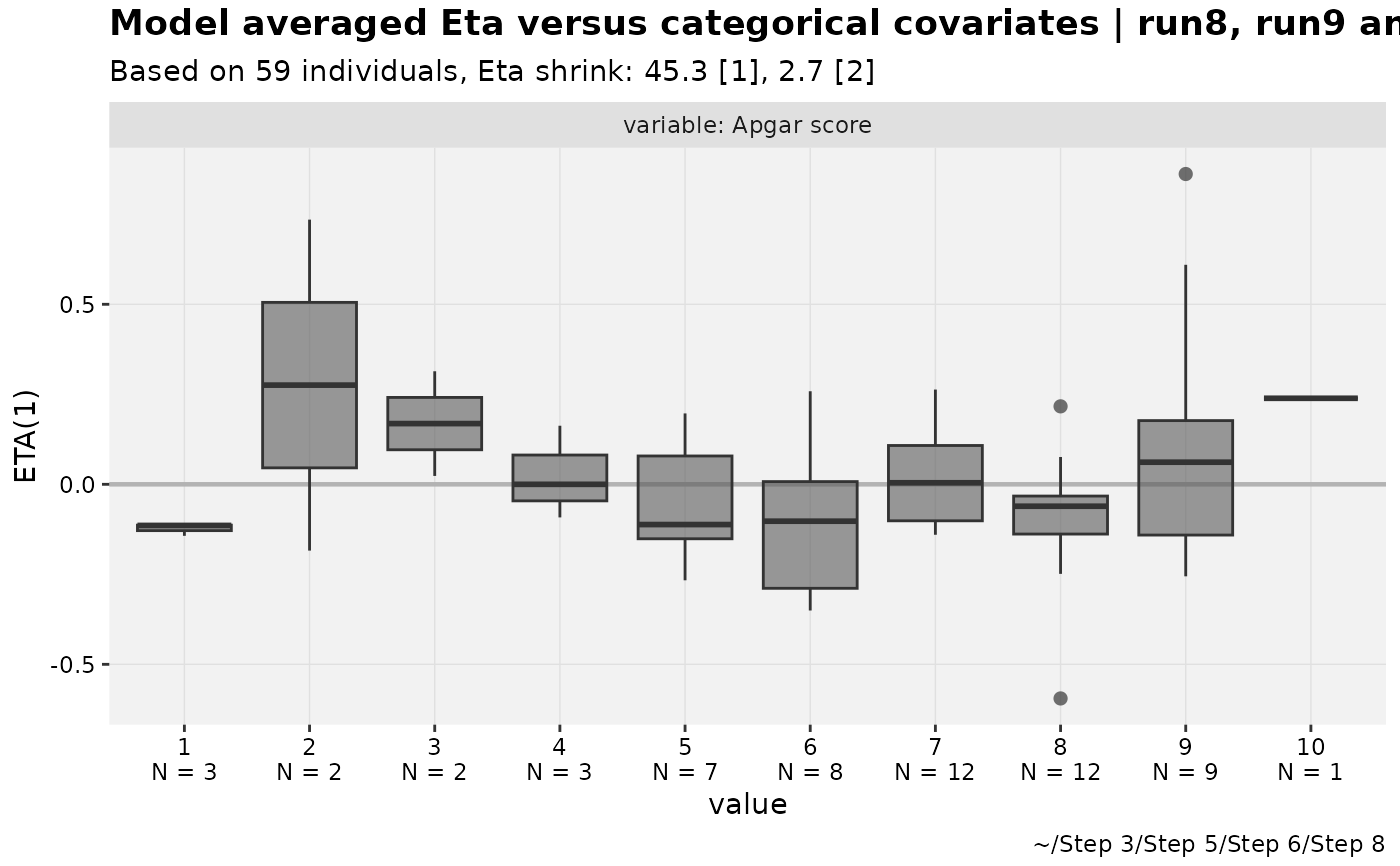
 # Model averaged ETA covariates
pheno_set %>%
plotfun_modavg(run8,run9,run10, auto_backfill = TRUE,
avg_by_type = "eta",.fun = eta_vs_catcov,
# Note quoting
.funargs = list(etavar=quote(ETA1)))
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Tidying data by ID, TIME, AMT, WT, MDV ... and 31 more variables
# Model averaged ETA covariates
pheno_set %>%
plotfun_modavg(run8,run9,run10, auto_backfill = TRUE,
avg_by_type = "eta",.fun = eta_vs_catcov,
# Note quoting
.funargs = list(etavar=quote(ETA1)))
#> Using data from $prob no.1
#> Removing duplicated rows based on: ID
#> Tidying data by ID, TIME, AMT, WT, MDV ... and 31 more variables
 # }
# }
